Scientists Extend Mechanism for Cracking Biochemical Code
A team of researchers from UC San Diego and Johns Hopkins University completes first comprehensive study related to histone coding and gene expression
Published Date
By:
- Cynthia Dillon
Share This:
Article Content
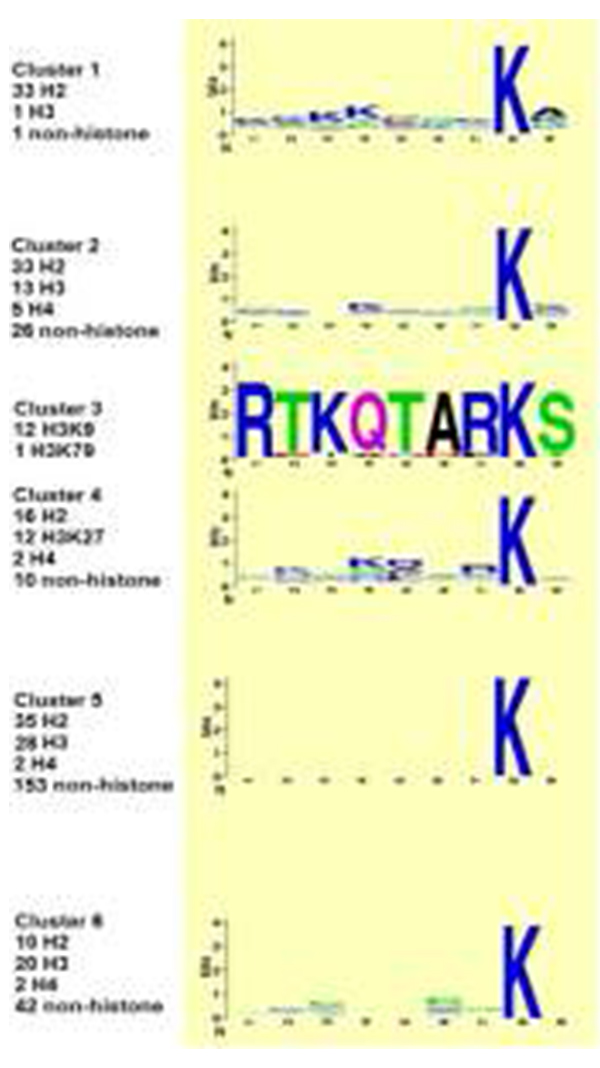
Graphic showing sequences for peptide clusters. Figure courtesy of Wei Wang
Since the time of ancient Egypt, humans have been making and breaking secret codes to retain and gain critical information. Human life itself is based upon a genetic code of DNA or RNA sequences which cells read and translate into proteins—the building blocks of life. Recent scientific discoveries have revealed the body’s mechanisms for transcribing DNA regulated by the “histone code”—different chemical marks on the tails of histone proteins, which are macromolecules within cell nuclei responsible for packaging and structuring DNA.
After eight years of study, a team of researchers from the University of California San Diego and Johns Hopkins University published new findings about how to read the body’s histone code in the Nov. 7 issue of Science Advances. The findings answer a key question in the dynamic research area of epigenetics—adding chemical tags to DNA and histone proteins to alter cell functions without changing DNA sequence. Understanding the fundamental principles of how epigenetic information is transduced in the cell eventually could lead to developing new drugs for fighting diseases like cancer.
In the research article titled “Deciphering and engineering chromodomain-methyllysine peptide recognition,” UC San Diego Professor of Chemistry and Biochemistry Wei Wang, postdoctoral scholars and his colleagues from the Departments of Pharmacology, Bioengineering, and Cellular and Molecular Medicine, as well as scientists from Johns Hopkins, provide a mechanistic explanation of how combinations of histone modifications could be read by certain proteins—“reader proteins”—leading to changes of gene expression and interpretation of information coded in the DNA genome.
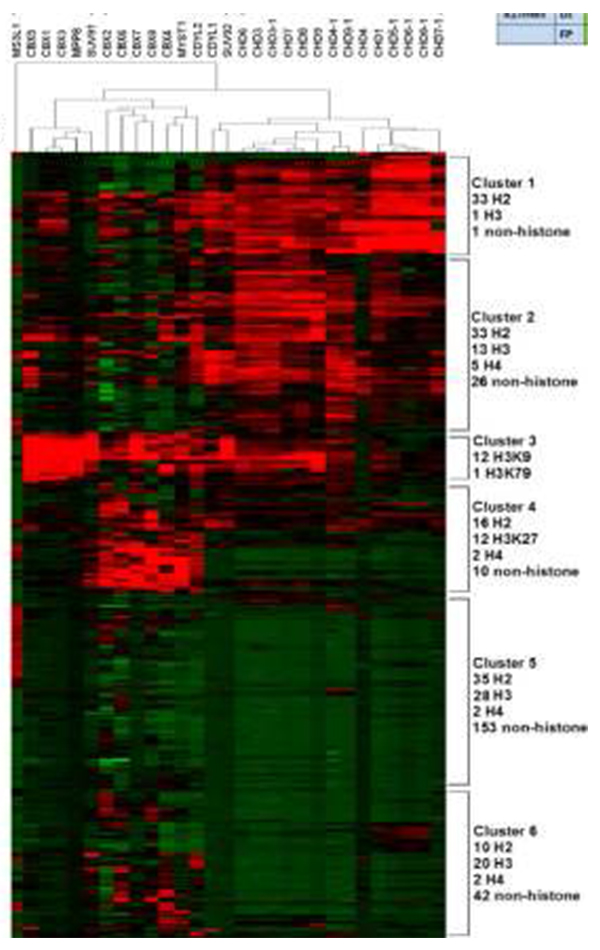
Chromodomain-peptide binding intensities on the microarray (shown as z scores; red, binding; green, nonbinding). Figure courtesy of Wei Wang
“We developed a model to understand how reader proteins see through different combinations of histone modifications, which interpret and transduce the information encoded in tagging histone proteins without changing the DNA sequence,” said Wang.
Applying a metaphor of makeup, Wang explained that if a person makes up his or her face, the makeup might change how the person looks, but he or she can still be identified as the same person. In the body, histone modifications can produce various combinations, like a person’s makeup, that can change the chemical properties of the histone proteins. But Wang and colleagues found that reader proteins recognize those same chemical properties, even if they resulted from different combinations of histone modifications. According to Wang, because the number of possible combinations of different chemical tags is huge, the histone code has not been well defined. The new findings, however, suggest a way to define the histone code, a computational model depicting the chemical properties of histone proteins. It is like seeing through a person’s makeup and revealing the person’s true identity.
“Based on the computational model, we are able to engineer the reader proteins to alter or enhance their binding to particular histone modifications,” explained Wang. “If reader proteins with certain mutations are generated, we can use them as imaging probes to monitor the dynamics of histone modifications in live cells. This is something that cannot be done using antibodies.”
The authors acknowledge C. Arrowsmith for providing the CBX1–8 constructs, which were modified for cloning. The research was supported by the National Institutes of Health (grants R01GM085188 and R35CA197622).
At UC San Diego, our research efforts are designed to change the world for the better—through new medicines, innovative technologies and more that will help address disease, global security, public policy, climate change and more.
Share This:
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



