Like Language, Genomes Are Encoded by Grammar Rules
Studies of marine invertebrate species reveal genome instructs development using logic and grammar
Story by:
Published Date
Article Content
The notochord serves a defining role in developing organisms across the animal kingdom. The flexible, rod-like structure supports the fragile emergence of growing embryos and serves as a key hub for signaling to ensure successful development of the nervous system and other cell types within our bodies.
University of California San Diego Assistant Professor Emma Farley and her research team are investigating how the right genes are expressed in the notochord to ensure successful development of embryos at this critical juncture. Enhancers ensure that genes are turned on at the right time and place. Errors in enhancers can lead to genes turning on in the wrong cell types, which can cause disease.

A study led by two Farley Lab graduate students, Ben Song and Michelle Ragsac, has found that notochord enhancers not only follow a precise pattern of gene expression, but the process is governed by rules that are similar to the ways in which grammar systematically structures our languages.
The findings, published in the journal Cell Reports, offer evidence proving how—similar to a string of words that must be grammatically arranged to create a functional sentence—our genomes contain specific features that must be organized in logical ways in order for notochord enhancers to do their jobs properly.
“Not every region in the genome that contains the same features is going to work the same way,” said Farley, a faculty member in the School of Medicine and School of Biological Sciences. “It’s how those features are arranged—similar to a functional sentence—in terms of their order, orientation and distance from each other.”
The researchers studied notochord enhancer activity in the marine invertebrate Ciona robusta, a model species for studying development and enhancers. Analyzing 90 genetic elements from Ciona embryos, the experiments produced some of the first-known evidence that enhancer grammar and logic rules are conserved across the spectrum of chordate species, organisms that belong to the phylum that includes vertebrates, tunicates and cephalochordates.
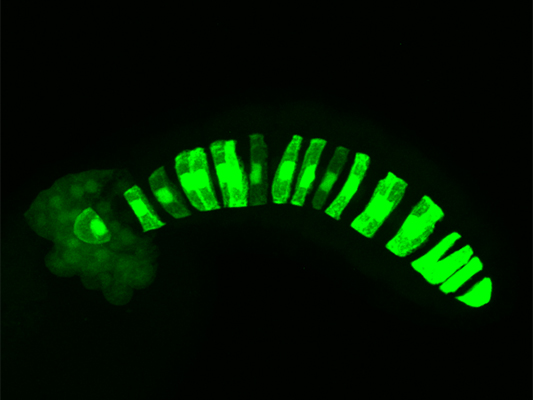
Green fluorescent protein, turned on by enhancers, or genomic switches, lights up the rod-like notochord of the marine invertebrate Ciona robusta.
“Collectively, our study finds that grammar is a key component of functional enhancers and we see signatures of this enhancer logic conserved across Ciona, mice and humans,” the authors note in the paper.
Farley and her research team are now seeking ways to use grammar signatures to read how the instructions for the development of organisms are encoded within genomes.
“We see signatures of this conserved grammar and logic across species, which suggests you could use grammar to understand how the instruction for making organisms is encoded in our DNA,” noted Farley. “It’s exciting to see grammar and the conservation of the grammar signature across organisms.”
Song, a Biological Sciences graduate student, and Ragsac, a Bioinformatics and Systems Biology graduate student, both graduated in Fall 2022 and are now working in biotech. Other authors include Krissie Tellez, Granton Jindal, Jessica Grudzien and Sophia Le.
Funding for the study was provided by the National Institutes of Health (T32 GM133351, T32 GM008666, T32HL007444 and DP2HG010013); National Science Foundation (2109907); a Hartwell Fellowship; American Heart Association (18POST34030077); and the UC San Diego Chancellor’s Research Excellence Scholars Program.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.




