What Makes A Bacterial Species Able to Cause Human Disease?
Global effort produces first cross-species genomic analysis of Leptospira, a bacterium that can cause disease – and death – in targeted mammals, including humans
Published Date
By:
- Scott LaFee
Share This:
Article Content
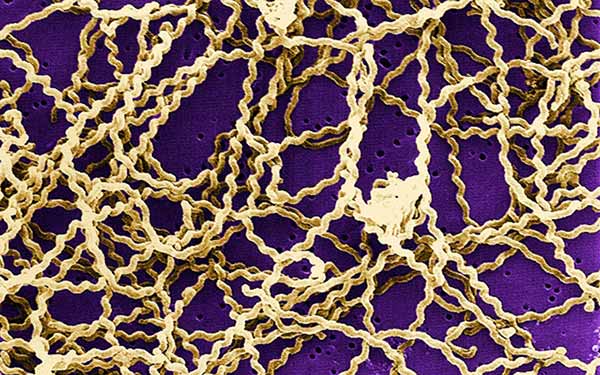
Scanning electron micrograph of Leptospira sp. Bacteria. Image courtesy of the Centers for Disease Control.
An international team of scientists, led by researchers at University of California, San Diego School of Medicine and the J. Craig Venter Institute (JCVI), have created the first comprehensive, cross-species genomic comparison of all 20 known species of Leptospira, a bacterial genus that can cause disease and death in livestock and other domesticated mammals, wildlife and humans.
The findings are published in the February 18 online issue of PLOS Neglected Tropical Diseases. The resulting analyses reveal novel adaptations and traits in infectious species of Leptospira that not only help illuminate its evolutionary history, but may also provide new preventive and treatment approaches. The data comes from representative Leptospira strains obtained from four international labs and isolated from 12 countries. The work involved researchers at Yale University, UCLA, Cornell University and institutions in Australia, France, England, The Netherlands, Canada, Uruguay, Brazil, Peru and Thailand.
"Leptospira is the most complex genus of bacteria that infects humans,” said Joseph M. Vinetz, MD, professor of medicine and director of the UC San Diego Center for Tropical Medicine and Travelers Health. “This work compares the complete genome sequences of all known species of Leptospira to discover which genes make this bacterium a pathogen. It provides a roadmap for future research, including finding new ways to diagnose infection and vaccine development.”
This project was initiated and led by first author Derrick Fouts, PhD, JCVI associate professor. “The bioinformatics tools have matured to the point that we can now find the proverbial ‘needle in a haystack,’ meaning we can now identify genes and pathways that are unique to pathogenic microbes, which will focus future research efforts on these key pathogen-specific features,” Fouts said.
JCVI president Karen E. Nelson, PhD, who helped coordinate the initial phase of the project points out that the success of this study was based, in part, on the dedication and excitement of the Leptospira research community. “The comprehensive nature of this study serves as a model for emerging infectious diseases.”
Some species of Leptospira are non-infectious, deriving nourishment from dead or decaying organic matter. Other species are infectious with different, sometimes dire, consequences. In infected rats and mice, for example, the bacterium colonizes in the kidneys, but does not produce disease symptoms. In livestock, such as cattle, sheep, horses and pigs, and companion animals, such as dogs, leptospirosis may cause acute kidney, liver and lung damage and provoke fetal loss.
Humans can become infected with leptospirosis in ways similar to animals, through direct contact or ingestion or inhalation of contaminated water or soil. Humans are also vulnerable through direct contact with the urine of infected animals.
In infected humans, the disease provokes a range of effects. It may result in asymptomatic infection, mild, flu-like symptoms or jaundice, or lead to severe damage of the liver, kidney or central nervous system – and death. Globally, leptospirosis is the most important bacterial disease transmitted from animals to humans, with more than 1 million cases annually and 60,000 deaths.
Examples of Leptospira pathogen-specific features identified in the PLOS study include:
- Discovery of proteins unique to pathogenic Leptospira that will accelerate vaccine development in humans, dogs, and livestock
- Key adaptations of infectious Leptospira to mammals, such as biosynthesis of sialic acid, a type of sugar that allows these organisms to hide within the body so as to avoid destruction and which allows disease to progress
- Infectious Leptospira evolutionarily acquired the ability to synthesize vitamin B12 from a simple amino acid precursor, which non-infectious Leptospira cannot
“One fascinating finding was discovering the CRISPR-Cas genetic machinery only in pathogenic Leptospira, but not in the intermediate or non-infectious groups of the genus. The evolutionary acquisition of novel CRISPR elements, which are only in pathogenic Leptospira, probably hastened adaptation to human infection. The significance of this observation remains to be explored,” said Vinetz.
CRISPR-Cas is a molecular immune system that confers resistance to foreign genetic elements, and provides a form of acquired immunity. Recent biotechnological advances have made it possible to alter the germline of many species, including mammals and disease-carrying mosquitoes, through CRISPR.
Vinetz said the next step will be to begin exploiting the genomic information “for vaccine and diagnostics development, experimental approaches to understanding mechanisms of disease pathogenesis and how Leptospira persist in the environment, all critical for developing new public health interventions aimed at reducing the global impact of this important but neglected zoonotic disease.”
Co-authors include Daniel H. Haft, J. Craig Venter Institute; Michael A. Matthias, David E. Berg, Jonathan M. Monk and Bernhard Palsson, UC San Diego; Haritha Adhikarla, Luciane Amorim-Santos, Albert I. Ko and Elsio A. Wunder Jr., Yale University and the Oswaldo Cruz Foundation; Ben Adler and Dieter Bulach, Monash University; Alejandro Buschiazzo, Institut Pasteur de Montevideo, Uruguay; Yung-Fu Chang, Cornell University; Renee L. Galloway, Centers for Disease Control; David A. Haake and James Matsunaga, VA Greater Los Angeles Healthcare System; Rudy Hartskeerl, Royal Tropical Institute, The Netherlands; Paul N. Levett, Saskatchewan Disease Control Laboratory, Canada; Ariel E. Mechaly and Mathieu Picardeau, Institut Pasteur; Ana L.T. Nascimento, Instituto Butantan, Brazil; Sharon J. Peacock, University of Cambridge; Jessica N. Ricaldi, Universidad Peruana Cayetano Heredia, Peru; Janjira Thaipandungpanit, Mahidol University, Thailand; and X. Frank Yang and Jun-Jie Zhang, Indiana University.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



