Going Viral
Computer modeling helps scientists understand airborne viruses.
Story by:
Media contact:
Published Date
Story by:
Media contact:
Share This:
Article Content
This story was published in the Fall 2023 issue of UC San Diego Magazine.
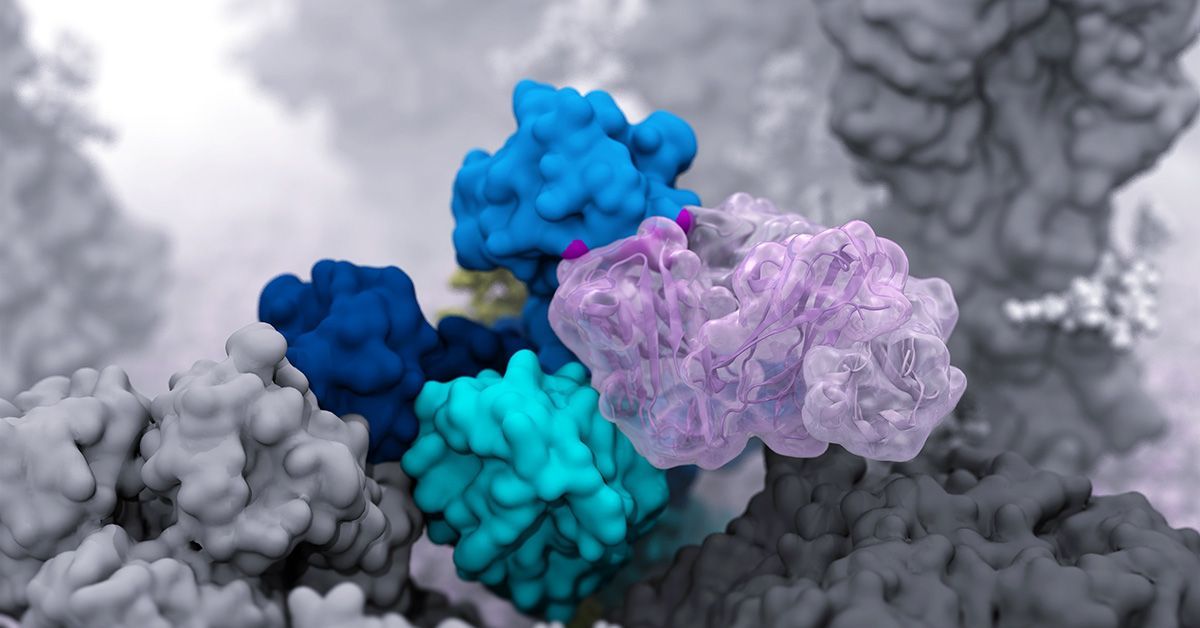
Can a computer save lives? If you ask Rommie Amaro, professor of molecular biology, the answer is yes. She and her team develop state-of-the-art computational techniques to investigate the structure, function and dynamics of complex biological systems, including viruses, such as SARS-CoV-2 and H1N1. Her simulations are giving epidemiologists and scientists new insights and tools in the battle against global diseases.
During the pandemic, the Amaro Lab created the first-ever atomic-level computer model of aerosolized SARS-CoV-2, the virus that causes COVID-19. Aerosols are tiny, smaller than the width of a human hair, and are produced simply by breathing and speaking. In this form, they can travel farther than other molecules and stay in the air for hours. “There is no microscope that allows people to see the particles in this much detail, but these computational models allow us to see what happens to the virus, like how it moves and how it stays infectious during flight,” says Amaro.
To better understand how the virus moves and lives inside aerosols, Amaro worked with Oak Ridge National Laboratory, using their Summit supercomputer to simulate the models. Summit is one of the few supercomputers in the world capable of performing these large-scale simulations, enabling researchers to see aerosols at a remarkable one billion atoms of detail. “No one had built a real system at this scale — there were only three computers in the world that could run these simulations, and it takes the whole machine to do it,” she says.

Rommie Amaro, professor of molecular biology at UC San Diego
Supercomputing simulations allowed Amaro’s team to develop animation footage of the virus in motion, offering a deeper understanding of the path of the COVID-19 virus in the human body.
Last year, Amaro’s lab went on to create an atomic-level computational model of the H1N1 influenza virus. The model revealed previously unknown “tilting” and “breathing” movements in two glycoproteins — vulnerabilities that may lead to more effective vaccines that don’t need to be reformulated each year.
The H1N1 virus simulation contains an enormous 160 million atoms-worth of detail. For this work, the lab used the Titan supercomputer, also at Oak Ridge National Lab and formerly one of the largest and fastest computers in the world.
“We have made our simulations available to any interested researchers,” says Amaro. “We’re interested in one thing, but other scientists will have different interests and they can mine this data — there are so many treasures to be discovered.”
Amaro’s work also supports UC San Diego’s new Meta-Institute for Airborne Disease in a Changing Climate, where she is a co-director of an interdisciplinary team of scientists focused on the impact of climate change on the air we breathe as well as the development of solutions to improve indoor and outdoor air quality globally.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.