Using Artificial Intelligence to Learn More about RAGE Receptors
High school student helps researchers further novel computational drug design tool
Published Date
By:
- Kimberly Mann Bruch
Share This:
Article Content
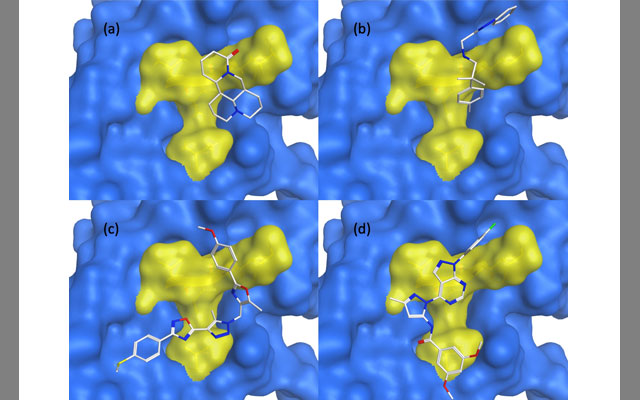
Receptor Advanced Glycation End-products (RAGEs) were first characterized in 1992 and thought to be linked to several chronic diseases including Alzheimer’s, diabetes, and congestive heart failure. They are the focus of a new study by researchers affiliated with the San Diego Supercomputer Center (SDSC) at UC San Diego, which describes the use of artificial intelligence machine learning tools to demonstrate a potential RAGE inhibitor that has better efficacy and fewer side effects.
The paper, published in January’s Physical Biology journal, is the result of studies by SDSC researchers Igor Tsigelny and Valentina Kouznetsova, as well as David Huang, a high school student participant in SDSC’s annual Research Experience for High School Students (REHS) summer internship program.
Glycation, which is the covalent attachment of a sugar, such as glucose, fructose, and their derivatives to protein, DNA or lipid amino groups, has been shown to be a key factor in AGE creation in the organism, according to the researchers. Previous inhibitors were developed mostly based on traditional pharmacophore design of compounds that block AGEs binding to RAGE.
Instead of relying solely upon pharmacophore-based research, Tsigelny, Kouznetsova, and Huang’s work used artificial intelligence (AI) to discover a new deep learning-based method of an inhibitor design. The new potential drug candidates were determined by the researchers to possibly be more effective, while having fewer negative side effects.
Fostering Student Involvement in Scientific Research
“The serious input to this publication was done by David Huang,” said Tsigelny. “We were thrilled to have an REHS student work with us on this published study, which demonstrates the potential of a broad family of deep-learning techniques on bioactivity predictions. David’s work was useful in allowing us to explain how inhibitors bind to RAGE – transmembrane receptors – and prevent binding of many advanced glycation end products (AGEs), which have been shown to enhance a large set of diseases.”
Huang has worked with Tsigelny and Kouznetsova on several other projects, including collaborating with other students to present a paper regarding treatment of diabetic cataracts.
“It is useful for our lab to have students working with us as they always present unique ideas that otherwise we might not think of,” said Tsigelny. “We have been really pleased with David’s work and look forward to watching his academic endeavors flourish.”
In addition to their positions at SDSC, Tsigelny and Kouznetsova are also affiliated with UC San Diego’s Moores Cancer Center. Tsigelny is also affiliated with the UC San Diego Department of Neurosciences and CureMatch Inc.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.