UC San Diego Leads Research that Earns Gordon Bell Special Prize
Computational Chemist Rommie Amaro and her research team win one of the most coveted awards in supercomputing for research related to COVID-19
Published Date
By:
- Rachel McDowell
- Cynthia Dillon
Share This:
Article Content
This year the Association for Computing Machinery (ACM) issued a special version of the Gordon Bell Prize, one of the most prestigious awards given for outstanding achievements in high performance computing. In recognition of COVID-19 research efforts, the Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research was presented Nov. 19 during the SC20 virtual conference. UC San Diego Computational Chemist and Biophysicist Rommie Amaro's team, including members of the Amaro Lab, Argonne National Laboratory (ANL) and Oak Ridge National Laboratory, won.
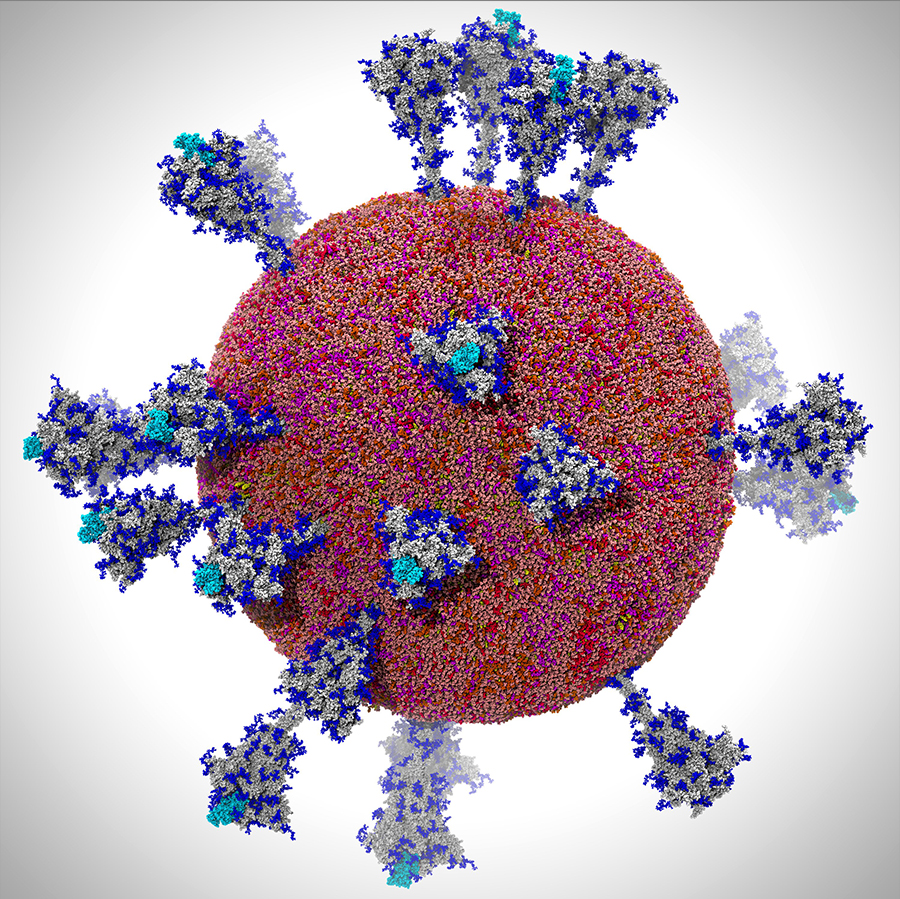
Multiscale modeling of SARS-CoV-2. All-atom model of the SARS-CoV-2 viral envelope (305 million atoms), including 24 spike proteins. Figure by UC San Diego’s Lorenzo Casalino, the Amaro Lab, and members of the research team.
The team led by Amaro and Arvind Ramanathan, computational biologist at ANL, has been exploring the movement of SARS-CoV-2’s spike protein to understand how it behaves and gains access to the human cell. Now, in a first-of-its-kind feat, the team has built a workflow based on artificial intelligence (AI) to more efficiently simulate the spike, and they have scaled the workflow to the Summit supercomputer to gain a deeper understanding of the spike’s mechanisms and accelerate the search for therapeutics or vaccines that might work to mitigate the virus.
“Experimental pictures give us a concept of what these things look like, but they can’t tell us the whole story,” Amaro said. “The only way we can do this is through simulations, and right now we are pushing the capabilities of molecular simulations to the limits of the computer architectures that we have on this earth. This is at the edge of possibilities of what people are capable of doing.”
The team first optimized the Nanoscale Molecular Dynamics (NAMD) and the Visual Molecular Dynamics (VMD) codes, which model the movements of atoms in time and space, on multiple smaller cluster systems: the Frontera supercomputer at the Texas Advanced Computing Center, the Comet system at the San Diego Supercomputer Center, and the ThetaGPU supercomputer at the Argonne Leadership Computing Facility (ALCF). The optimizations prepared them to run their full-scale simulations on the OLCF’s Summit. The OLCF and the ALCF are US Department of Energy (DOE) Office of Science User Facilities located at DOE’s Oak Ridge and Argonne national laboratories, respectively.
After code optimizations, the team was able to successfully scale NAMD to 24,576 of Summit’s NVIDIA V100 GPUs. The results of the team’s initial runs on Summit have led to discoveries of one of the mechanisms that the virus uses to evade detection as well as a characterization of interactions between the spike protein and the protein that the virus takes advantage of in human cells to gain entrance into them—the ACE2 receptor.

Professor of Chemistry and Biochemistry Rommie Amaro. Photo courtesy of Rommie Amaro.
“This is one of the first biological systems of the virus that we can learn from to drive scientific discovery,” Amaro said. “Our methods of computing allow us to get down to actually see detailed intricacies of this virus that are useful for understanding not only how it behaves but also its vulnerabilities, from a vaccine development standpoint, and a drug targeting perspective.”
Because one set of the calculations generated a whopping 200 terabytes of data, the team used AI to identify the intrinsic features from the simulations and break down the information to help them interpret what was happening. By layering the experimental data and the simulation data and combining it with their AI-based approach, the researchers were able to capture the virus and its mechanisms in unprecedented detail. The team is also integrating the NAMD code into their workflow pipeline to fully automate the transition from simulation to AI for data processing without gaps.
“We never thought we could use our machine-learning tools at this scale,” Ramanathan said. “Using these AI-based approaches on Summit have helped accelerate the process of truly understanding the motion of these complex systems.”
This research was supported by the Exascale Computing Project; the DOE National Virtual Biotechnology Laboratory, with funding provided by the Coronavirus CARES Act; and the COVID-19 HPC Consortium.
Related Publication: Lorenzo Casalino, Abigail Dommer, Zied Gaieb, Emilia P. Barros, Terra Sztain, Surl-Hee Ahn, Anda Trifan, Alexander Brace, Anthony Bogetti, Heng Ma, Hyungro Lee, Matteo Turilli, Syma Khalid, Lillian Chong, Carlos Simmerling, David J. Hardy, Julio D. C. Maia, James C. Phillips, Thorsten Kurth, Abraham Stern, Lei Huang, John McCalpin, Mahidhar Tatineni, Tom Gibbs, John E. Stone, Shantenu Jha, Arvind Ramanathan, and Rommie E. Amaro. “AI-Driven Multiscale Simulations Illuminate Mechanisms of SARS-CoV-2 Spike Dynamics.” In Proceedings of SC20, Virtual Event, November 16-19, 2020.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



