The Antibiotic-Resistant Bacteria Problem Is Getting Worse. Phage Could Be a Solution.
Innovative undergraduate research class results in scientific publication
Published Date
Article Content
The situation is dire and depressing: you’ve woken up in the middle of the night, your ear throbbing and swollen. Not another ear infection! The solution seems simple and straightforward: go for the antibiotics to treat the bacteria, right? In many cases, that might work. But if the bacteria is antibiotic resistant, what’s next? Try this on for size — a prescription cocktail chock-full of viruses.
It’s called phage therapy, and scientists in the University of California San Diego School of Biological Sciences in Professor Joe Pogliano’s laboratory are advancing research into the bacteriophages. These viruses are harmless to humans but specifically target certain bacteria, including important pathogens like Escherichia coli, that are at the heart of this promising treatment.

UC San Diego graduate student Amy Prichard and UC San Diego undergraduate student Jina Lee led a study in Pogliano’s lab to understand the mechanism by which a certain family of bacteriophages protect themselves against the bacteria they’re infecting. The paper, published in Cell Reports, identifies the core set of genes that encode the proteins behind this method.
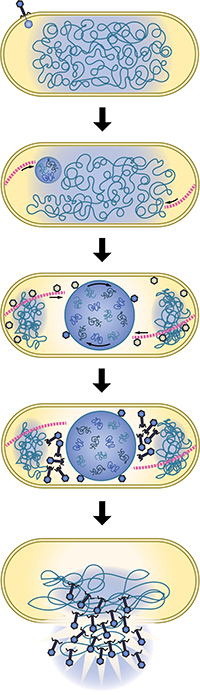
But Prichard and Lee aren’t the only authors — there are many more of them, in fact — and the vast majority of them (28!) were first-year UC San Diego biology students.
The students are advancing research on special bacteriophages Pogliano’s group studies. Normally, when bacteriophages infect bacteria, bacteria can use several self-defense mechanisms to chop up the viral DNA, like restriction enzymes and CRISPR-Cas systems. His team found, however, that some viruses put together a nucleus-like structure that protects the DNA from the bacteria’s defense systems. But nuclei, which house DNA in eukaryotic cells, are uncommon in prokaryotic cells, and thus their finding warranted new classification. They named these types of bacteriophages “chimalliviruses'' after chimallin, the major component of the phage nucleus.
“It used to be thought that a virus would come in, it would replicate in random positions all over the place, make progeny phage and lyse the bacterium,” said Pogliano. “We never knew these kinds of complex structures could form in a bacterial cell, let alone be encoded by a virus.”
The discovery of the phage nucleus spurred many questions: what was the core recipe — the genes behind the phage nucleus? Were there more virus families that exhibited the same behavior?
Prichard set out in search, examining many diverse bacteriophages that encode chimallin, the protein that forms the phage nucleus. She eventually settled on vB_EamM_RAY, a bacteriophage that infects Erwinia, a pathogenic bacteria that infects apple and pear trees.
But, like many other scientists’ research, work on RAY and what goes into the phage nucleus shuttered when the pandemic hit.
'We were doing real research'
Prichard wasn’t the only one stuck because of the pandemic. Pogliano too was mired in a quandary: the class he was supposed to teach in fall 2020 was forced online. He teaches the Phage Genomics Research Initiative, a two-part lab class that takes place over two quarters, where students go out into the field to isolate new bacteriophages in Fall quarter and return in Spring quarter to characterize them. The class is unique because the students publish their results from the class. That means students get to dip their hands immediately into research.
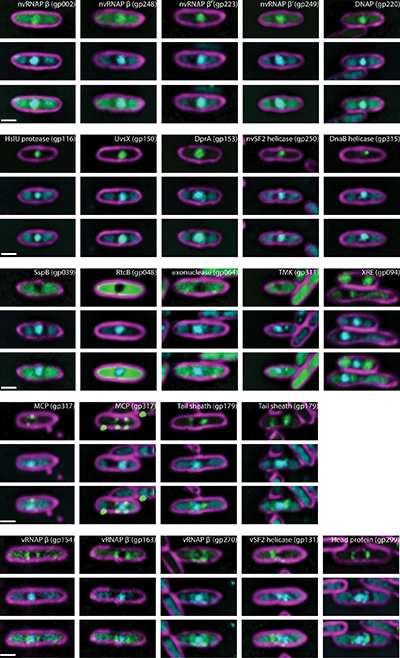
Images of fluorescently labeled RAY proteins designed by the first-year students in the Phage Genomics Research Initiative Class. Credit: Amy Prichard
“In a traditional classroom, if you did the experiment, and you didn’t measure the correct weight of the molecules, you get a B minus. Here, if the experiment doesn’t work, you do it again until you figure out why it’s not working so you get a result and isolate a virus,” Pogliano said. “In fact, there are some class periods where we learn more when the experiments fail than when the experiments work. We discuss it as a group. Why did our experiments fail? Oh, this thing was contaminated. Well, why is it contaminated?” added Pogliano, stressing the importance of persistence in research.
Just one big problem was in the way, however — the class was now online. So Pogliano and Prichard put their heads together and came up with a new approach that solved both of their problems: why not have the students use bioinformatics to learn about the proteins that RAY encodes?
During the class, the students analyzed a subset of RAY’s proteins to understand their functions and the families that they belonged to. Each person eventually hammered in on one protein to study, which became their own project over the course of the year.
“It felt hands-on as opposed to a lecture class. We were doing real research. I was looking at a protein in RAY and nobody else in the world knew more about that protein than I did,” said Annika Sy, a student who took the class.

Zoom class of the Phage Genomics Research Initiative students, including Professor Rachel Dutton and graduate instructional apprentices Tara Spencer and Amy Prichard, Spring 2021. Credit: Rachel Dutton
Then followed the students’ next hurdle: writing up their results at the end of spring quarter. It sounded like a herculean group project nightmare — twenty-eight pairs of hands typing away at a single Google Doc — but it culminated in a first draft of a scientific paper.
“It was a fun and very kind of different experience for undergraduates, especially for freshmen.” Pogliano said.
While the students went home satisfied with their work, Prichard was convinced there could be more to RAY and its proteins. A look at the phylogenetic tree the students built — with the help of Professor Rachel Dutton, the instructor for the Spring quarter of the class — showed that there were a handful of clear cut-offs where distinct bacteriophage groups shared different amounts of conserved RAY proteins the students studied. Some groups made intuitive sense; they shared almost all of the homologs and were RAY’s close genetic relatives. But another group that appeared to share numerous genes with RAY seemed odd as it did not share the phage nucleus replication strategy. Upon closer inspection, eight of the conserved homologs were RNA polymerases.
“These bacteriophages share a lot of genes [that encode for things] like DNA polymerases. This group [of phages] bothered me — there’s this cutoff where these two genes are differently conserved here and there. We need to analyze the whole genome of this bacteriophage to see what’s conserved, not just 30 of these proteins,” Prichard said, wondering if all of the phages that encode chimallin had more in common than just the genes shared with the group in green.
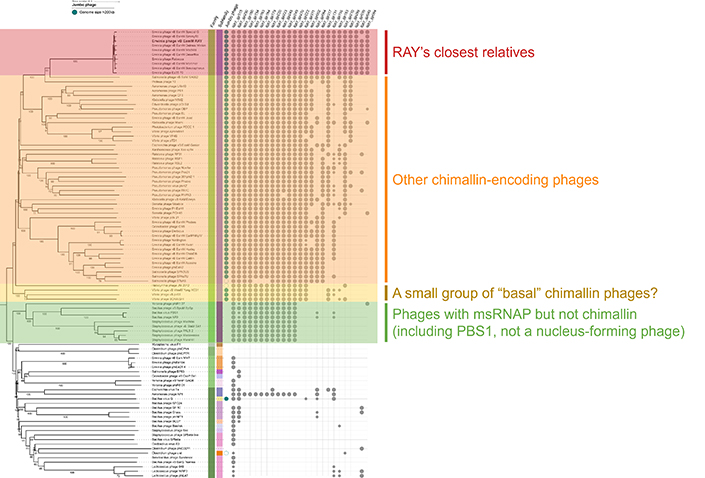
A phylogenetic tree (left) of the bacteriophages with RAY in bold, and the genes in RAY (top) studied by the students. Each dot represents the presence of a similar gene in the phage across from it. Credit: Rachel Dutton and Amy Prichard
Enter Jina Lee, a Phage Genomics Research Initiative alumni and a member of Pogliano’s lab, who could provide Prichard with those answers. A bioinformatics student, Lee broadened the project’s scope by tapping into her computational skills to examine the genes of all bacteriophages and comparing the genes found in phages predicted to form a phage nucleus. By specifically examining phages encoding homologs of the chimallin protein, the two of them hoped to find a clearer explanation for the discrepancies that they had found.
Lee’s analysis proved fruitful. “I noticed that some proteins are very well conserved and some proteins are not conserved at all. They’re very specific to RAY and some Erwinia phages,” Lee said. “I found out all of the phages had homologs to this one RAY protein.”
Finding that there were several blocks of genes that were inherited together (seven in total, containing 68 genes) demonstrated a tangible, genetic connection between the bacteriophages that were being studied. More importantly, these genes were ones involved in the many facets of nucleus formation, genome replication, gene expression and formation of virion particles. As such, the group had characterized the “core genome” of nucleus-forming bacteriophages.
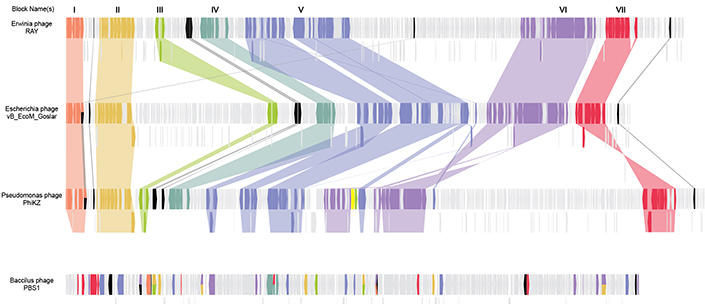
The genomes of RAY and two other chimallin-encoding phages are shown here. Blocks of genes that perform similar functions are highlighted across all three genomes, representing seven different conserved groups. The bottom phage does not encode chimallin, and its genome has much less similarity. Credit: Jina Lee.
The final puzzle pieces were set: all Prichard, Lee and Pogliano had to do now was to name their discovery. Since the common factor linking these phages together had been chimallin, the major protein component of the phage nucleus, the two of them aptly titled this family Chimalliviridae.
'Science is a community'
This kind of understanding of how bacteriophages work is a key interest of The Center for Innovative Phage Applications and Therapeutics (IPATH), a research institute founded at UC San Diego in 2018. IPATH, a leading center on phage therapy, experiments with bacteriophage to treat bacterial diseases that are resistant to antibiotics.
IPATH’s work has already had exciting progress. Earlier this year, several brands of artificial eye drops were linked to outbreaks of Pseudomonas aeruginosa, which can cause deadly eye infections if not treated quickly. The researchers there had isolated bacteriophage that targeted Pseudomonas aeruginosa and used the phage to kill the bacteria in samples of the eye drops sent by the CDC.
In addition, the phage class has already had one case of taking the research to the bedside. One freshman in a prior offering of the class discovered a bacteriophage, Fionnbharth, that ended up being used by Graham Hatfull’s laboratory at the University of Pittsburgh to treat two patients infected with Mycobacterium. Dr. Hatfull has been spearheading the approach of using students to find phage that can be used for phage therapy. In fact, RAY and other Erwinia phages were originally isolated by undergrads at Brigham Young University in 2018 under Julianne Grose.
“Science is a community. We’re going to share and compare phage with each other. This is a great example of how that worked,” Pogliano said. “Their students isolated the phage and effectively sent them to us. Our students worked on them along with phage students from other classes and then put together a publication with extremely nice discoveries.”
“The other thing we’ve been telling students since 2008 is: why should you work on phage? Well, maybe one day – phage therapy.” Pogliano added. “15 years later, it’s happening everyday.”
The study’s full author list includes Amy Prichard, Jina Lee, Thomas G. Laughlin, Amber Lee, Kyle P. Thomas, Annika E. Sy, Tara Spencer, Aileen Asavavimol, Allison Cafferata, Mia Cameron, Nicholas Chiu, Demyan Davydov, Isha Desai, Gabriel Diaz, Melissa Guereca, Kiley Hearst, Leyi Huang, Emily Jacobs, Annika Johnson, Samuel Kahn, Ryan Koch, Adamari Martinez, Meliné Norquist, Tyler Pau, Gino Prasad, Katrina Saam, Milan Sandhu, Angel Jose Sarabia, Siena Schumaker, Aaron Sonin, Ariya Uyeno, Alison Zhao, Kevin D. Corbett, Kit Pogliano, Justin Meyer, Julianne H. Grose, Elizabeth Villa, Rachel Dutton and Joe Pogliano.
Funding for this research came from the National Institutes of Health (grant R01 GM129245), the National Science Foundation (grant DBI 1920374), and the Howard Hughes Medical Institute Emerging Pathogens Initiative.
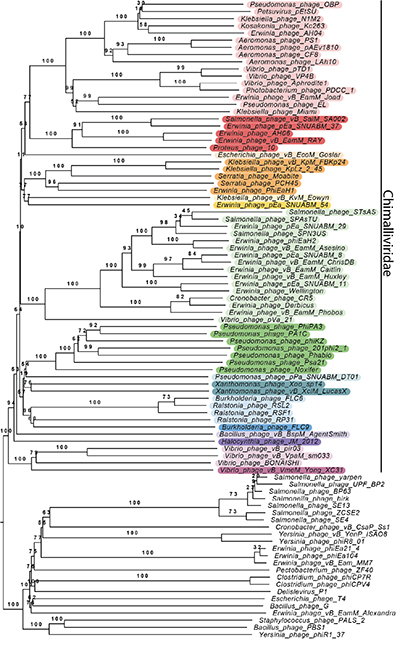
A phylogenetic tree showing the many bacteriophages grouped together by genetic similarity. The chimalliviruses are colored in rainbow colors. RAY and its group of Erwinia phages are highlighted in red. Credit: Amy Prichard
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



