By:
- Jorge Salazar
- Cynthia Dillon
Published Date
By:
- Jorge Salazar
- Cynthia Dillon
Share This:
Sugar-Coating Disguise Allows for Coronavirus Infection
According to Mary Poppins, a spoonful of sugar helps the medicine go down. In the case of coronavirus, a cloak of sugar helps the virus infect. This sugary-coating disguise, made of molecules called glycans, tricks the human immune system into identifying the microbe as harmless. The resulting recognition failure keeps the body from generating the defensive antibodies needed to destroy the invading coronavirus.

Rommie Amaro, professor of chemistry and biochemistry, UC San Diego
Using the National Science Foundation-funded Frontera supercomputer at the Texas Advanced Computing Center (TACC), Professor of Chemistry and Biochemistry Rommie Amaro—along with her UC San Diego colleagues and researchers from Maynooth University in Dublin, Ireland, led by Elisa Fadda—has uncovered the atomic makeup of the coronavirus's sugary cloak. The simulation and modeling reveal that glycans also prime the coronavirus for infection by changing the shape of its spike protein. Scientists hope this basic research will add to the arsenal of knowledge needed to defeat the COVID-19 virus.
“The more we know about it, the more of its abilities that we're going to be able to go after and potentially take out,” Amaro said. “It is of such great importance that we learn as much as we can about the virus. And then hopefully we can translate those understandings into things that will be useful either in the clinic or the streets; for example, if we're trying to reduce transmission for what we know now about aerosols and wearing masks. All these things will be part of it. Basic research has a huge role to play in the war against COVID-19. And I'm happy to be a part of it. It's a strength that we have Frontera and TACC in our arsenal.”
Glycans coat each of the 65-odd spike proteins that adorn the coronavirus. The sugar-like molecules account for about 40 percent of the spike protein by weight. The spike proteins are critical to cell infection because they lock onto the cell surface, giving the virus entry into the cell.
Amaro, along with her UC San Diego colleagues Lorenzo Casalino, Zied Gaieb, Abigail Dommer, Emilia Barros and Bryn Taylor, explained that even to make an initial connection, one of the pieces of the spike protein in its receptor binding domain has to lift up. It is one of the things Frontera—part of the COVID-19 HPC Consortium along with San Diego Supercomputer Center at UC San Diego—helped reveal: that in the open conformation, there are two glycans that basically prop up the spike protein.
“That was really surprising to see. It's one of the major results of our study. It suggests that the role of glycans in this case is going beyond shielding to potentially having these chemical groups actually being involved in the dynamics of the spike protein,” said Amaro, a corresponding author of the study published online June 12, 2020, by bioRxiv,org, a preprint repository.
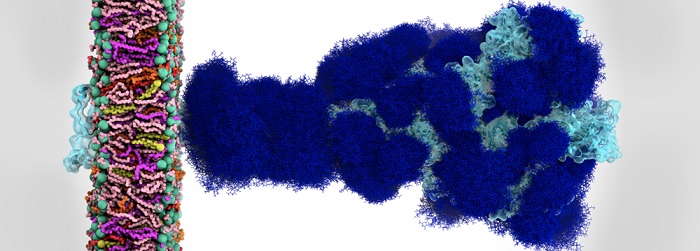
Glycan shield in SARS-CoV-2 spike. (A) Molecular representation of the Open. Glycans at several frames (every 20 ns) are represented with blue lines, and the receptor binding domain within chain A is highlighted with a cyan transparent surface. (B-C) Plot of the surface area covered by glycan shielding at multiple probe radii from 1.4 Å (water molecule) to 15 Å for the head (B) and stalk (C). The area of the protein covered by the glycans is depicted in blue, while the grey line is the accessible area of the protein without glycans. Highlighted in green is the area that remains accessible in the presence of glycans, which is also graphically depicted on the structure in the panels located above the plots. Credit: Lorenzo Casalino (UC San Diego), et al.
“When that receptor binding domain lifts up into the open conformation, it actually lifts the important bits of the protein up over the glycan shield,” Amaro said, adding that this contrasts with the closed conformation, where the shield covers the spike protein. “Our analysis gives a potential reason why it does have to undergo these conformational changes, because if it just stays in the down position those glycans are basically going to block the binding from actually happening,” she said, adding that the shifts in the conformations of the glycans triggered changes in the spike protein structure.
Amaro compared the action of the glycan to pulling the trigger of a gun. “When that bit of the spike goes up, the finger is on the trigger of the infection machinery. That's when it's in its most dangerous mode—it is locked and loaded,” Amaro said. “When it gets like that, all it has to do is come up against an ACE2 receptor in the human cell, and then it's going to bind super tightly and the cell is basically infected.”
The research team used computational methods to build data-centric models of the SARS-CoV-2 virus, and then used computer simulations to explore different scientific questions about the virus. They started with various experimental datasets that revealed the structure of the virus. This included cryo-EM structures from the Jason McLellan Lab of The University of Texas at Austin; and from the lab of David Veesler at the University of Washington.
“Their structures are really amazing because they give researchers a picture of what these important molecular machines actually look like,” Amaro said.
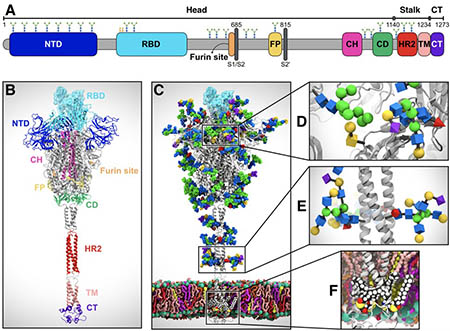
SARS-CoV-2 virus spike protein system overview. (A) Sequence of the full-length spike protein contains the N-terminal domain (NTD), the receptor binding domain (RBD), the furin cleavage site, the fusion peptide (FP), the central helix (CH), the connecting domain (CD), the heptad repeat 2 (HR2) domain, the transmembrane domain (TD) and the cytoplasmic tail (CT). (B) Assembly of head, stalk, and cytoplasmic tail (CT) sections into a full-length model of the spike protein (C) Equilibrated, fully glycosylated and palmitoylated model of the Open system. (C-E) Magnified view of the N-/O- glycans (C, D) and S-palmitoylation of the cytoplasmic tail (E). Image by Lorenzo Casalino, et al.
Unfortunately, even the most powerful microscopes on Earth still can't resolve movement of the protein at the atomic scale.
“What we do with computers is that we take the beautiful and wonderful and important data that they give us, but then we use methods to build in missing bits of information,” Amaro said. “What people really want to know — for example, vaccine and drug developers — are the vulnerabilities that are present in this shield.”
The computer simulations allowed Amaro and colleagues to create a cohesive picture of the spike protein that includes the glycans.
“The reason why the computer resources at TACC are so important is that we can't understand what these glycans look like if we don't use simulation,” Amaro said.
In order to animate the dynamics of the 1.7 million atom system under study, a lot of computing power was needed, said Amaro.
“That's really where Frontera has been fantastic, because we need to sample relatively long dynamics, microsecond to millisecond timescales, to understand how this protein is actually working. We've been able to do that with Frontera and the COVID-19 HPC Consortium,” Amaro said. “Now we're trying to share our data with as many people as we can, because people want a dynamical understanding of what's happening—not only with other academic groups, but also with different pharmaceutical and biotech companies that are conducting neutralizing antibody development,” she said, adding that basic research is making a difference in winning the war against the SARS-Co-V-2 virus.
This research was supported by NIH (GM132826), NSF (RAPID MCB-2032054), an award from the RCSA Research Corp., a UC San Diego Moore's Cancer Center 2020 SARS-COV-2 seed grant, the Visible Molecular Cell Consortium and the Irish Research Council.
Share This:
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



