Researchers Uncover Achilles Heel of Antibiotic-Resistant Bacteria
As drug resistance surges, scientists discover a promising new way to control the spread of this public health crisis
Story by:
Published Date
Article Content
Recent estimates indicate that deadly antibiotic-resistant infections will rapidly escalate over the next quarter century. More than 1 million people died from drug-resistant infections each year from 1990 to 2021, a recent study reported, with new projections surging to nearly 2 million deaths each year by 2050.
In an effort to counteract this public health crisis, scientists are looking for new solutions inside the intricate mechanics of bacterial infection. A study led by researchers at the University of California San Diego has discovered a vulnerability within strains of bacteria that are antibiotic resistant.
Working with labs at Arizona State University and the Universitat Pompeu Fabra (Spain), Professor Gürol Süel and colleagues in UC San Diego’s School of Biological Sciences investigated the antibiotic resistance of the bacterium Bacillus subtilis. Their research was motivated by the question of why mutant variants of bacteria do not proliferate and take over the population once they have developed an antibiotic-resistant advantage. With an upper hand over other bacteria lacking similar antibiotic resistance, such bacteria should become dominant. Yet they are not. Why?
The answer, reported in the journal Science Advances, is that antibiotic resistance comes at a cost. While antibiotic resistance provides some advantages for the bacteria to survive, the team discovered that it’s also linked with a physiological limitation that hinders potential dominance. This fact, the researchers note, potentially could be exploited to stop the spread of antibiotic resistance.
“We discovered an Achilles heel of antibiotic resistant bacteria,” said Süel, a member of the Department of Molecular Biology at UC San Diego. “We can take advantage of this cost to suppress the establishment of antibiotic resistance without drugs or harmful chemicals.”
Spontaneous mutations of DNA arise in all living cells, including those within bacteria. Some of these mutations lead to antibiotic resistance. Süel and his colleagues focused on physiological mechanisms related to ribosomes, the micro machines within cells that play a key role in synthesizing proteins and translating genetic codes.
All cells rely on charged ions such as magnesium ions to survive. Ribosomes are dependent upon magnesium ions since this metal cation helps stabilize their structure and function. However, atomic-scale modeling during the new research found that mutant ribosome variants that bestow antibiotic resistance excessively compete for magnesium ions with adenosine triphosphate (ATP) molecules, which provide energy to drive living cells. Mathematical models further showed that this results in a ribosome versus ATP tug-of-war over a limited supply of magnesium in the cell.
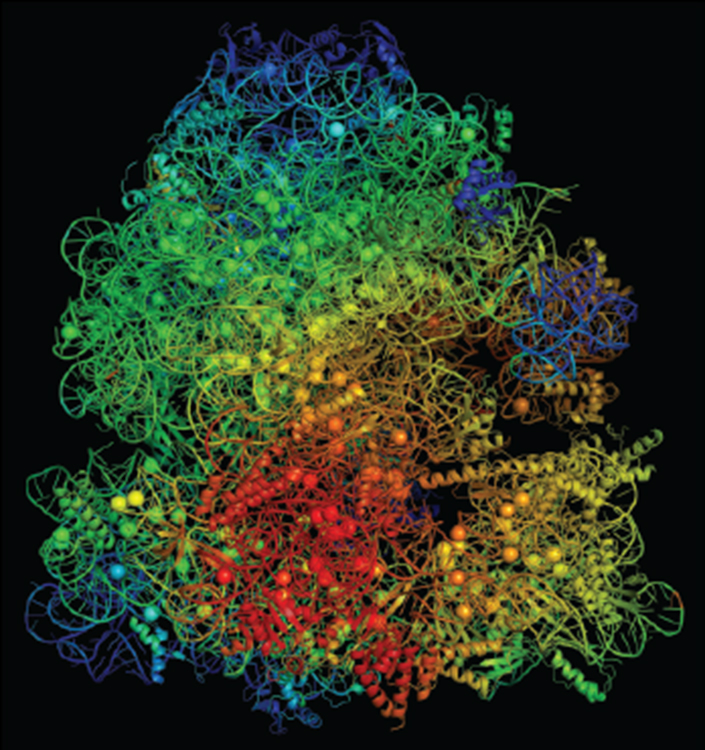
A model structure of a ribosome with color-coded flexibility indicators; red highlights ribosome regions that become more flexible, while blue depicts more rigid areas.
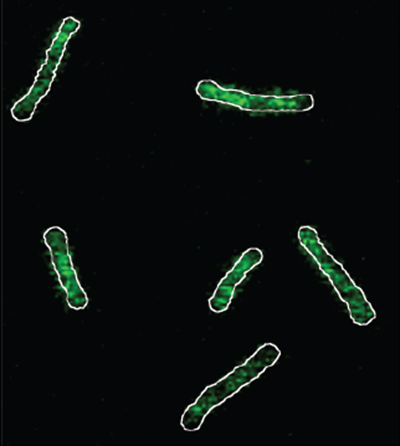
Image depicts the outlines of bacterial cells with green fluorescence highlighting a lack of magnesium.
Studying a ribosome variant within Bacillus subtilis called “L22,” the researchers found that competition for magnesium hinders the growth of L22 more than a normal “wild type” ribosome that is not resistant to antibiotics. Hence the competition levies a physiological toll linked to mutant bacteria with resistance.
“While we often think of antibiotic resistance as a major benefit for bacteria to survive, we found that the ability to cope with magnesium limitation in their environment is more important for bacterial proliferation,” said Süel.
This newly discovered weakness can now be used as a target to counteract antibiotic resistance without the use of drugs or toxic chemicals. For example, it may be possible to chelate magnesium ions from bacterial environments, which should selectively inhibit resistant strains without impacting the wild type bacteria that may be beneficial to our health. “We show that through a better understanding of the molecular and physiological properties of antibiotic-resistant bacteria, we can find novel ways to control them without the use of drugs,” said Süel.
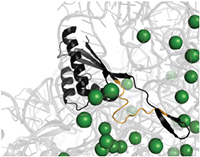
A magnified view of the ribosome variant within Bacillus subtilis known as L22 (black) and surrounding magnesium ions (green dots).
In October Süel and colleagues at the University of Chicago announced a separate approach to combating the antibacterial-resistant bacteria health crisis. Their development of a bioelectronic device that taps into the natural electrical activity of certain bacteria found on our skin paves the way for another drug-free approach to managing infections. The advancement was proven to reduce the harmful effects of Staphylococcus epidermidis, a common bacterium known for causing hospital-acquired infections and contributing to antibiotic resistance. In both studies the researchers used charged ions to control bacteria.
“We are running out of effective antibiotics and their rampant use over the decades has resulted in antibiotics being spread across the globe, from the arctic to the oceans and our groundwater,” said Süel. “Drug-free alternatives to treating bacterial infections are needed and our two most recent studies show how we can indeed achieve drug-free control over antibiotic resistant bacteria.”
The authors of the new study were: Eun Chae Moon, Tushar Modi, Dong-yeon Lee, Danis Yangaliev, Jordi Garcia-Ojalvo, S. Banu Ozkan and Gürol Süel.
The research was funded by the National Institute of General Medical Sciences (grant R35 GM139645); Army Research Office (grant W911NF-22-1-0107 and W911NF-1-0361); Bill and Melinda Gates Foundation (INV-067331); Spanish Ministry of Science, Innovation and Universities and FEDER project (PID2021-127311NB-I00 and CEX2018-000792-M); Generalitat de Catalunya ICREA Academia program; National Science Foundation Division of Molecular and Cellular Biosciences (award 1715591) and the Gordon and Betty Moore Foundation.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



