Novel Molecular Dynamics Captures Atomic-level Detail of CRISPR-Cas9 Activity
SDSC’s ‘Comet’ helps break the “millisecond barrier” for complex biological simulations
Published Date
By:
- Warren Froelich
Share This:
Article Content
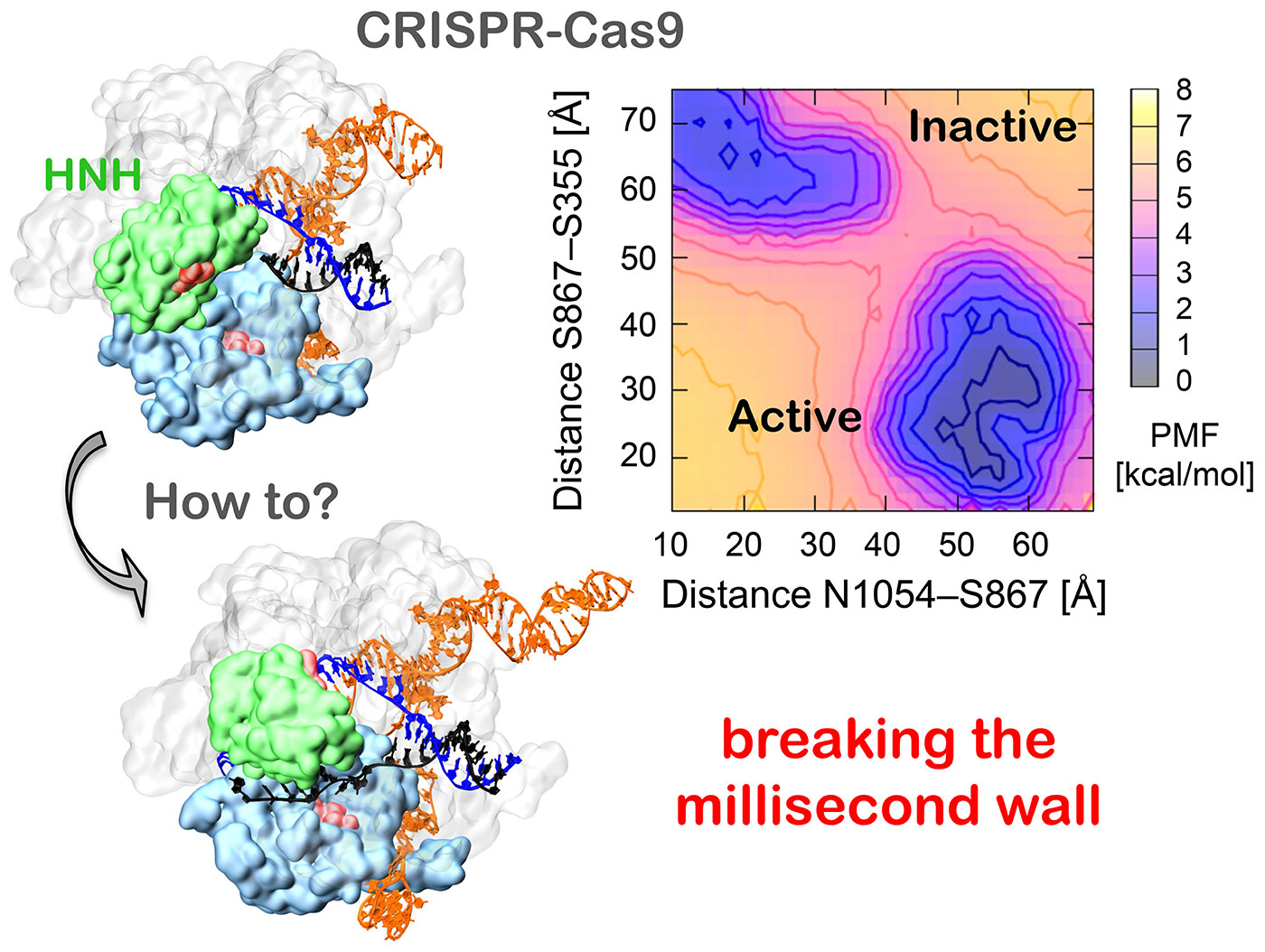
CRISPR-Cas9 conformational activation, as revealed from Gaussian accelerated Molecular Dynamics (GaMD) simulations. GaMD reveals the conformational change of the HNH domain (green) from its inactive configuration (top) to the active state (bottom). This conformational change, which occurs along milliseconds, is captured with atomic-level precision via the use of advanced simulation methods (i.e. GaMD) and state-of-the-art supercomputing resources. Credit: Giulia Palermo, McCammon Lab, UC San Diego
Using a novel molecular dynamics method capable of capturing the motion of gyrating proteins at time intervals up to one thousand times greater than previous efforts, a team led by UC San Diego researchers has identified, for the first time, the myriad structural changes that activate and drive CRISPR-Cas9, the innovative gene-splicing technology that’s transforming the field of genetic engineering.
By shedding light on the biophysical details governing the mechanics of CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats) activity, the study provides a fundamental framework for designing a more efficient and accurate genome-splicing technology that doesn’t yield “off-target” DNA breaks currently frustrating the potential of the CRISPR-Cas9- system, particularly for clinical uses.
“Although the CRISPR-Cas9 system is rapidly revolutionizing life sciences toward a facile genome editing technology, structural and mechanistic details underlying its function have remained unknown,” said Giulia Palermo, a postdoctoral scholar with the UC San Diego Department of Pharmacology and lead author of the study, published in the June 26 early edition of the Proceedings of the National Academy of Sciences (PNAS).
“In particular, we want to design a system that doesn’t cause ‘off-target’ effects or non-selective cleavage of DNA sequences, that can now create unwanted collateral damage,” added J. Andrew McCammon, the Joseph E. Mayer Chair of Theoretical Chemistry at UC San Diego, a Howard Hughes Medical Institute Investigator, and the study’s principal investigator.
As its name implies, CRISPR-Cas9 is a dual entity with dual functions. The first consists of a short RNA guide molecule, part of which matches a target DNA sequence; the second is a Cas9 enzyme that recognizes and slices the DNA in a precise spot, whose location or address is post-marked by a nucleotide sequence called a protospace adjacent motif, or PAM. The result is an RNA-DNA hybrid with a displaced non-target DNA strand.
Dubbed Science magazine’s “breakthrough of the year” in 2015, researchers are looking to apply the technology to a variety of uses, with hopes of treating diseases through gene therapy or driving advances in areas from crop engineering to the production of biofuels.
But the transformative tool isn’t without its flaws. Studies have revealed that the RNA guide used to direct the cleaving enzyme to its target can sometimes go astray, landing on other DNA strands with similar but not identical sequences. The result is “off-target” mutations, severely limiting the technology for clinical applications.
Thus, a worldwide search has begun for a clearer picture of the system’s inner molecular workings with the goal of engineering a new version without the current system’s inherent flaws.
Although extensive studies of the CRISPR-Cas9 system, including X-ray crystallography and cryoelectron microscopy (cryoEM) have revealed detailed views of the system’s structural and biological activity, the dynamics of Cas9 and its step-by-step acrobatics with nucleic acids during its merger and cleavage of DNA have remained fuzzy.
Capturing Protein Dynamics via Comet
Last year, researchers from the McCammon Lab at UC San Diego used the Comet supercomputer at SDSC to perform atomistic molecular dynamics – a method that captures a more complete vision of the myriad shapes and conformations that a target protein molecule may go through – at petascale speeds (one quadrillion arithmetic calculations per second).
The resulting simulations, published in the September 8 issue of ACS Central Science, identified some of the key factors underling the complex structural changes taking place during the merger and preparation for cleaving the target DNA strand.
Though explaining some of the fundamental biophysics of the system, these relatively short-lived simulations – conducted upward of multi-microseconds – could not clarify the conformational transitions arising from the protein’s resting state to the catalytically active Cas9 state.
To overcome this limitation, the researchers turned to a novel methodology called Gaussian accelerated molecular dynamics or GaMD, created by Yinglong Miao in McCammon’s lab, a process that lengthens the observational time-scale from microseconds (millionths of a second) to milliseconds (thousandths of a second). This advance allows researchers to simulate more complex biophysical transitions, including protein folding and ligand binding, while also capturing complex structural transitions.
“The fact that GaMD breaks the millisecond barrier is of great importance, because it gives us the opportunity to observe biological processes that are out of reach of experiments with atomic-level resolution,” said Miao, a co-author of the PNAS study.
Turning to Comet, Once Again
Using GaMD, the researchers once again returned primarily to Comet – in addition to SDSC’s Triton Shared Computing Cluster (TSCC) and Bridges at the Pittsburgh Supercomputing Center – to perform CRISPR-Cas9 simulations at the lengthened time-scale. Supercomputer allocations for Comet and Bridges were made available by the Extreme Science and Engineering Discovery Environment (XSEDE), one of the most advanced collections of integrated digital resources and services in the world.
The results provided “on-the-fly” atomic-level detail of conformational changes that take place in the CRISPR-Cas9 system, leading the Cas9 protein from its resting state to its final RNA-bound catalytic state. Until now, structural information about active Cas9 has been missing, in spite of extensive efforts by structural biologists worldwide.
“We showed that, upon DNA binding, the conformation dynamics of the active domain site (HNH) of Cas9 triggers the formation of the active state, explaining how the HNH domain exerts a conformational control domain over DNA cleavage,” said Palermo. “Knowledge of the catalytic state greatly advances and helps in the effort of engineering CRISPR-Cas9 systems with improved specificity.”
“In light of the fact that CRISPR-Cas9 still suffers from lack of on-target specificity, this is a great advance,” added McCammon, also a distinguished professor of pharmacology at UC San Diego.
As their next step, the research team is expecting to use this latest data to design a Cas9 protein that overcomes the limitations of current CRISPR-Cas9 technology.
“We hope this effort will make CRISPR-Cas9 applicable without risk of aberrant genome expressions,” said Palermo.
Also participating in the research were Ross Walker, previously with SDSC; and Martin Jinek, University of Zurich. Support for this study came from an advanced post-doctoral grant to Palermo from the Swiss National Science Foundation; grants to the McCammon Lab from the National Institutes of Health, National Science Foundation, Howard Hughes Medical Institute, and National Biomedical Computational Resource among others; research fellowships to Ross Walker from Intel and NVIDIA; and computing time from XSEDE with the grants MCA93S013 and MCB 160059.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



