New Class of Anti-infective Drugs May Overcome Antibiotic Resistance
SDSC’s ‘Gordon’ Supercomputer used to test compounds for uncoupler activities
By:
- Warren Froelich
Media Contact:
- Jan Zverina - jzverina@sdsc.edu
- Warren R. Froelich - froelich@sdsc.edu
Published Date
By:
- Warren Froelich
Share This:
Article Content
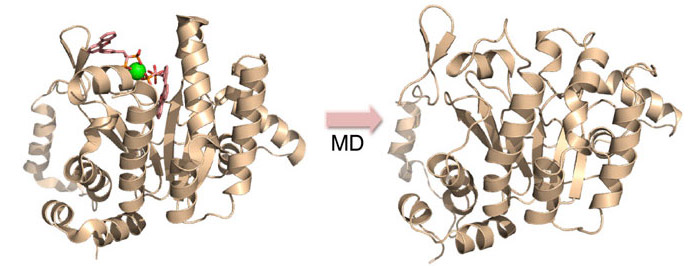
Vacquinol and its analogs as inhibitors for Rv3378c activity and bacterial cell growth. This image from Figure 5 in the published paper shows MD simulation/in silico screening approach used to identify Rv3378c inhibitors, with the X-ray structure (Left) and MD snapshot (Right). Image by J. Andrew McCammon, UC San Diego.
Antibiotic resistance has been called one of the world’s most pressing health problems, with some studies suggesting that, absent major improvements in drug discovery, by 2050 more individuals will die from drug-resistant bacterial infections than cancer.
To help stem this nightmare scenario, a team of researchers – with the aid of the Gordon supercomputer at the San Diego Supercomputer Center based at UC San Diego – has identified a class of possible antibiotics with the potential to disable previously drug-resistant bacteria.
In essence, these new agents were found to attack the bacteria along two fronts: its external lipid cellular wall and its internal factory responsible for generating cellular energy in the form of adenosine triphosphate or ATP.
“The exciting thing here is the discovery of a new dual mode of action of antibiotics – one that inhibits enzymes that bacteria like the one causing tuberculosis need to survive; and collapsing the electrical potential across the bacterial cell membrane, which blocks synthesis of the cellular energy molecule ATP,” said contributing author J. Andrew McCammon, the Joseph E. Mayer Chair Professor of Theoretical Chemistry and Distinguished Professor of Pharmacology at UC San Diego; and an Investigator with the Howard Hughes Medical Institute.
In a study published in the December 7 online edition of the Proceedings of the National Academy of Sciences (PNAS), the researchers showed that some FDA-approved anti-infective drugs act in part by disabling a cell’s ability to synthesize the production of ATP. These so-called “uncouplers” work by collapsing a cell’s “proton motive force”, a reservoir of protons that powers protein turbines used to synthesize ATP, much the way the flow of water through mechanized turbines generates electricity.
The researchers further discovered that a few newly approved anti-tuberculosis drugs worked not only as uncoupler agents, but they also targeted enzymes that maintain a bacteria cell’s outer membrane.
“This multiple targeting is expected to be of importance in overcoming the development of drug resistance because targeting membrane physical properties is expected to be less susceptible to the development of resistance,” said McCammon.
Using molecular dynamics-based in silico screening, the researchers sought to identify other compounds with dual-targeting capabilities. Among other things, the team discovered that the brain cancer drug lead Vacquinol was one such compound, which was of particular interest since the drug also has direct killing activity against the tuberculosis bacteria.
With the aid of the data-intensive supercomputer Gordon, the team tested a series of compounds for uncoupler activity in hopes of finding more potent analogs with activity against the tuberculosis bacteria and S. (staphlococcus) aureus infections. Subsequent experiments revealed several agents with uncoupling capabilities, in addition to others with at least dual targeting capabilities. For example, the new S. aureus growth inhibitor clomiphene, known to target cell wall biosynthesis, also demonstrated uncoupler activity.
“Pure uncouplers (without enzyme targets) are generally not expected to be good drug leads,” said McCammon. “But the results indicate that screening for combined enzyme inhibition plus uncoupler activity could lead to new antibiotic leads,” said McCammon.
Other researchers participating in the study include: Xinxin Feng, Wei Zhu, Lici A. Schurig-Briccio, Jikun Li, Yang Wang, Noman Baig, Tianhui Zhou, Boo Kyung Kim, Robert B. Gennis and Eric Oldfield (University of Illinois); Steffen Lindert (Ohio State University); Carolyn Shoen and Michael Cynamon (Veterans Affairs Medical Center, Syracuse, N.Y.); and Reese Hitchings and Dean C. Crick (Colorado State University). Feng, McCammon, and Oldfield co-authored the study. The work at UC San Diego was supported in part by the National Science Foundation, National Institutes of Health, the Howard Hughes Medical Institute, and the National Biomedical Computation Resource.
Share This:
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



