New Imaging Tool Advances Study of Lipid Biology
The imaging tool has created new opportunities to study the roles that specific lipid subtypes play in health, aging and in disease
Story by:
Published Date
Article Content
From fruit flies to humans, there are many, many different types and subtypes of lipids operating at the same time within any living organism. For example, the plasma portion of human blood is home to 600 different types of lipids. While we know that lipid molecules play myriad different roles in health, aging and disease; researchers currently struggle to uncover the fine details of these roles – details which could unlock cures, extend the human healthspan, and solve mysteries of aging.
One big challenge is connected to the fact that multiple subtypes of lipids are often found within the very same cells. At the same time, there is no ideal tool for identifying and tracking the activity of specific lipids within individual cells. While, there is a long list of existing techniques that are used to try to answer some of the toughest questions about the roles of specific lipid subtypes in individual cells and tissues – all current techniques have drawbacks.
Now, a study led by bioengineers at the University of California San Diego marks a significant step forward in this critical area of lipid research. The new work was published on February 21, 2024 in the journal Nature Communications. In this paper, the research team presents what they believe is the first method for distinguishing multiple lipid subtypes in cells and tissue samples by using nondestructive label-free optical imaging methods.
Introducing the New Imaging Platform
The new light-based imaging tool is called a hyperspectral imaging platform or PRM-SRS. The new capabilities of the PRM-SRS platform are due, in part, to the fact that the imaging platform integrates a Penalized Reference Matching (PRM) algorithm with Stimulated Raman Scattering (SRS) microscopy.
Use of this platform could profoundly deepen researchers’ ability to understand the roles that different lipid subtypes are playing in a wide range of cells and tissues. This is particularly relevant because traditional imaging methods often fall short in capturing the intricate spatial distributions and metabolic dynamics of lipid subtypes, hampering efforts to unravel their significance in aging and various diseases.
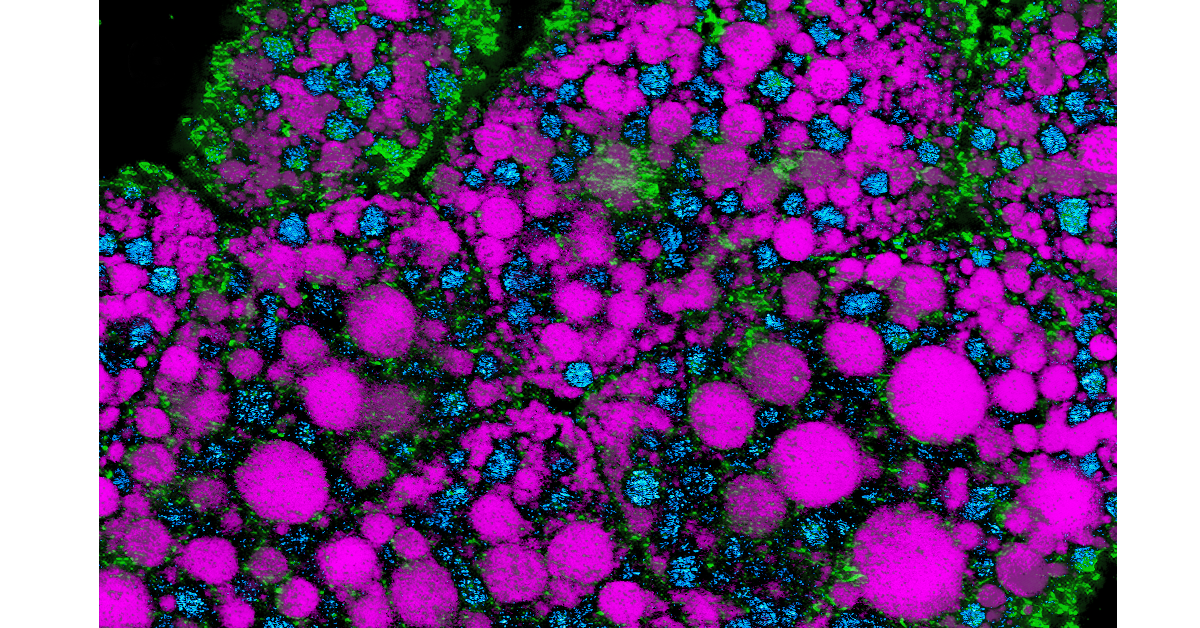
Demonstration of the new imaging platform
"Our PRM-SRS platform represents a paradigm shift in lipid imaging, offering unparalleled capabilities in distinguishing lipid subtypes within complex biological environments," said UC San Diego bioengineering professor Lingyan Shi, the senior corresponding author. “As far as we know, we are presenting the first method for distinguishing multiple lipid subtypes in cells and tissue samples by using nondestructive label-free optical imaging methods.” Shi was selected as an Alfred P. Sloan Research Fellow in 2023.
As described in the Nature Communications paper, the researchers harnessed PRM-SRS to visualize and identify distinct lipid subtypes across different organs and species including: high density lipoprotein particles in human kidneys; cholesterol-rich granule cells in mouse hippocampus; and subcellular distributions of two lipids (sphingosine and cardiolipin) in the human brain. In these demonstration cases, the PRM-SRS imaging platform unveiled unprecedented insights with enhanced chemical specificity (which is used to identify lipid subtypes) and subcellular resolution.
Highlights from the New Study
Multiplexed visualization: Unlike conventional methods, PRM-SRS enables the simultaneous visualization of multiple lipid subtypes from single label-free hyperspectral imaging (HSI) sets, expanding the scope of lipid research.
Diagnostic potential: Preliminary analyses of human kidney tissue samples suggest a potential application of PRM-SRS in diagnosing and prognosing renal diseases, such as offering non-invasive insights into dyslipidemia-associated conditions.
Neurological insights: By mapping lipid distributions in mouse and human brain tissues, PRM-SRS sheds light on the role of lipid metabolism in neurological disorders, paving the way for targeted therapeutic interventions.
Future directions: With its versatility and ease of implementation, PRM-SRS holds promise for diverse applications, from high-throughput studies to deep learning-enhanced imaging techniques, fostering a new era of multiplex cell and tissue imaging.
Paper Information
Multi-molecular hyperspectral PRM-SRS microscopy in Nature Communications / https://doi.org/10.1038/s41467-024-45576-6
This project is led by researchers in the Shu Chien-Gene Lay Department of Bioengineering at the UC San Diego Jacobs School of Engineering. This collaborative project includes important contributions from researchers from Northwestern University School of Medicine, the UC Irvine School of Medicine, Duke University School of Medicine, Duke University’s School of Medicine and Department of Biomedical Engineering, and Washington University in St. Louis.
Paper authors: Wenxu Zhang, Yajuan Li, Anthony A. Fung, Zhi Li, Hongje Jang, Honghao Zha, Xiaoping Chen, Fangyuan Gao, Jane Y. Wu, Huaxin Sheng, Junjie Yao, Dorota Skowronska-Krawczyk, Sanjay Jain, Lingyan Shi
The researchers report no conflict of interest.
Funding
UC San Diego Startup funds, NIH R01GM149976, NIH U01AI167892, NIH 5R01NS111039,NIH R21NS125395, NIH U54DK134301, NIHU54 HL165443, NIH U54CA132378, and a Hellman Fellow Award. We are grateful for the support of the Washington University Kidney Translational Research Center (KTRC) for kidney samples and the HuBMAP grant U54HL145608. We thank Dr. E. Bigio and Dr. M.-M. Mesulam from Mesulam Center for Cognitive Neurology and Alzheimer’s Disease (MCCNAD) for providing the de-identified autopsy brain samples; and MCCNAD is supported by NIH P30 AG013854. Work done in D.S.K. laboratory is supported by NEI P30 grant, P30EY034070-01 and in part by an unrestricted grant from Research to Prevent Blindness awarded to the Gavin Herbert Eye Institute.
Human BioMolecular Atlas Program
This paper is also collected by the NIH HubMAP program’s Nature series publication collection here: Human BioMolecular Atlas Program (nature.com)
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.