Looking Beyond DNA to See Cancer with New Clarity
Mapping how mutated proteins interact reveals previously unknown drug targets
Published Date
By:
- Heather Buschman, PhD
Share This:
Article Content
Researchers at University of California San Diego and University of California San Francisco have mapped out how hundreds of mutations involved in two types of cancer affect the activity of discrete groups of proteins that are the ultimate actors behind the disease. The work points the way to identifying new precision treatments that may skirt side effects common with much current chemotherapy.
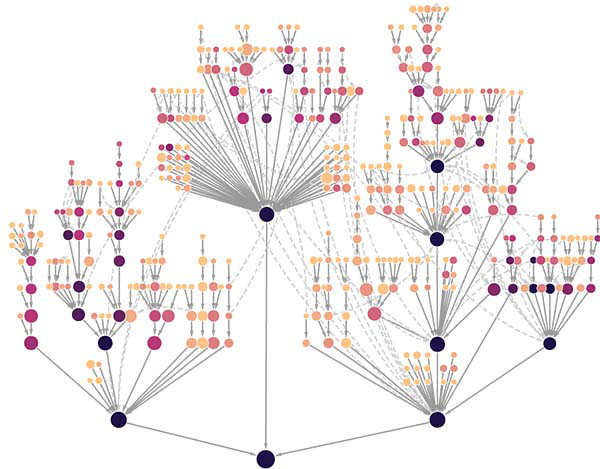
Hierarchy of cancer protein systems: Each node represents a protein system carrying out cellular functions such as mobility or immune signaling. Nodes farther out on the branches represent systems with few proteins and highly specialized processes, while those closer to the root have many proteins and correspond to generalized processes. Darker colored systems, and their subsystems, are under selection in more tumor types.
The effort, dubbed Cancer Cell Mapping Initiative (CCMI), is led by Trey Ideker, PhD, professor at UC San Diego School of Medicine and Moores Cancer Center, and Nevan Krogan, PhD, director of the Quantitative Biosciences Institute at UCSF, who are co-senior authors on a set of three related studies that describe the map. The papers appear in the October 1, 2021 online issue of Science.
“The bottom line is that we’re elevating the conversation about cancer from individual genes to whole protein complexes,” Ideker said. “For years, different groups have been discovering more and more mutations that are involved in cancers, but in so many different genes that scientists can’t make sense of it all. Now we’re able to explain these mutations at the next level — by looking at how the different gene mutations in different patients actually have the same downstream effects on the same protein machines.
“This is the first map of cancer from the protein complex lens.”
Mapping protein mutations
DNA contains the instructions for building proteins, which then interact with other proteins, almost always in large groups called complexes. These protein complexes, in turn, make up most of the machinery of cells, dictating basic cell functions like feeding, growth and whether the cell develops into cancer. If the underlying DNA has a mutation, the resulting protein machines often will as well.
In cancers, a subset of genes is commonly mutated, Krogan said, and each of these genes can be mutated in hundreds of different ways. In addition, the function of a particular protein may be different in different types of cells, so a mutation in a breast cancer cell might have different effects on protein complexes than that same mutation in a cell in the throat.
CCMI’s goal was to map the constellation of protein complexes formed by approximately 60 proteins commonly involved in either breast cancer or cancers of the head and neck, and to see what each looked like in healthy cells. Alongside that effort, they created maps of how protein complexes are affected by hundreds of different gene mutations in two cancerous cell lines.
“This is an exciting advance that not only provides a treasure trove of new protein-protein interactions, but also the computational tools to robustly analyze the data and put it in a meaningful context for others to use,” said Shannon K. Hughes, PhD, deputy director of the Division of Cancer Biology at the National Cancer Institute, part of the National Institutes of Health, which funds CCMI. “The methodology can be expanded to other tumor types and other diseases, which is very exciting. Importantly, these large-scale, systems-level mapping endeavors require a strong collaborative team, which has clearly been demonstrated within the CCMI.”
Expanding precision medicine
Currently, physicians look for a small number of mutated genes as biomarkers to decide whether or not to prescribe a particular drug. For instance, patients with breast cancer who have an alteration in their HER2 gene are given the medication Herceptin because that’s what will work best for them.
“The problem is that there are still only a few genes that work in this way, providing reliable biomarkers that are clearly actionable with an FDA-approved drug,” Ideker said. “Our studies provide a new definition of biomarkers based not on single genes or proteins but on large, multi-protein complexes.”
Because each protein complex incorporates mutations from a larger collection of genes, it is typically relevant to more patients, Ideker said. For example, XRCC5 is a DNA-repair gene altered in just 2 percent of colon cancers, which limits the usefulness of this biomarker. Now, however, researchers can look at CCMI’s new map of cancer protein complexes and see that XRCC5 is part of a 15-protein assembly altered in 14 percent of patients, and that these patients are typically very resistant to standard therapies.

Trey Ideker, PhD, professor at UC San Diego School of Medicine and Moores Cancer Center.
“There are many examples like this in our map,” Ideker said. “The clear next step is to help scientists and physicians evaluate them for use in the clinic. This is one of the main reasons we have worked really hard to make the map easily accessible on the web.”
“Indeed, by targeting simultaneously multiple components of these ‘oncogenic networks,’ our collaborative studies will pave the way for the development of more effective combination cancer therapies, while preventing treatment resistance,” said co-author J. Silvio Gutkind, PhD, chair of the Department of Pharmacology at UC San Diego School of Medicine and associate director of basic science and co-director of the Head and Neck Cancer Center at Moores Cancer Center. “These studies in breast and oral cancer can now be expanded to most human malignancies.”
The most powerful aspect of these extensive protein interaction maps is that they can shed the same light on many other conditions, Krogan said. For example, the team is also at work on similar studies of protein interactions in psychiatric and neurodegenerative disorders and infectious diseases.
Collaboration is key
The team sees the CCMI collaboration as the real source of strength behind the approach.
“We're not only making connections between different genes and proteins, but also between different people and different disciplines,” Krogan said. “Those collaborations have built up an infrastructure that allows them to integrate an array of types of information and push the boundaries of what’s possible in applying data science to complex diseases.
“We’re in the perfect position to take advantage of this revolution on every level. I couldn’t be more excited than I am right now. We can do such damage to cancer.”
“Interpretation of cancer mutations using a multiscale map of protein systems”
Co-authors include: Fan Zheng, Marcus R. Kelly, Dana J. Ramms, Marissa L. Heintschel, John J. Lee, Keiichiro Ono, Michael Chen, Erica Silva, Sophie Liu, Jing Chen, Christopher Churas, Nicholas Wilson, Anton Kratz, Rudolf Pillich, Devin N. Patel, Jisoo Park, Brent Kuenzi, Michael K. Yu, Katherine Licon, Dexter Pratt, Jason F. Kreisberg, Stephanie I. Fraley, J. Silvio Gutkind, Trey Ideker, UC San Diego; Kai Tao, Xiaolin Nan, Oregon Health and Science University; Beril Tutuncuoglu, Helene Foussard, Minkyu Kim, Danielle L. Swaney, Nevan J. Krogan, UC San Francisco and J. David Gladstone Institutes; Kari A. Herrington, UC San Francisco.
“A protein network map of head and neck cancer reveals PIK3CA mutant drug sensitivity”
Co-authors include: Danielle L. Swaney, Margaret Soucheray, Kyumin Kim, Michael McGregor, Helene Foussard, John Von Dollen, Mehdi Bouhaddou, David Jimenez-Morales, Kirsten Obernier, Minkyu Kim, Nevan J. Krogan, UC San Francisco and J. David Gladstone Institutes; Dana J. Ramms, Zhiyong Wang, Jisoo Park, Yusuke Goto, Fan Zheng, Jason F. Kreisberg, J. Silvio Gutkind, Trey Ideker, UC San Diego; Neil Bhola, Yan Zeng, Kari A. Herrington, Rachel O’Keefe, Nan Jin, Nathan K. VanLandingham, Daniel E. Johnson, Natalia Jura, and Jennifer R. Grandis, UC San Francisco.
“A protein interaction landscape of breast cancer”
Co-authors include: Minkyu Kim, Mehdi Bouhaddou, Kyumin Kim, Ajda Rojc, Maya Modak, Margaret Soucheray, Michael J. McGregor, Erica Stevenson, Beril Tutuncuoglu, Kuei-Ho Chen, Danielle L. Swaney, Nevan Krogan, UC San Francisco and J. David Gladstone Institutes; Jisoo Park, Fan Zheng, Jason F. Kreisberg, Trey Ideker, UC San Diego; Patrick O’Leary, Denise Wolf, Dominique Mitchell, Kari A. Herrington, Denise P. Muñoz, Morgan E. Diolaiti, John D. Gordan, Jean-Philippe Coppé, Laura van’t Veer, Alan Ashworth, UC San Francisco; Tzeh Keong Foo, and Bing Xia, Rutgers Cancer Institute of New Jersey.
Funding for this research came, in part, from the National Institutes of Health (U54CA209891, U54CA209988, U24CA184427, P41GM103504, R01HG009979, R01GM132322, R01DE026870, R01DE028289, R01CA247551, R01GM109176, R01GM143427, R01CA138804, R35CA231998, R50CA243885, F32CA239336, F32CA239333, 5F30CA236404-02, K00CA212456, S10OD026929), National Science Foundation (grant MCB-1651855), American Cancer Society (RSG-21-033-01-CSM), V foundation, BRCA foundation, Martha and Bruce Atwater Breast Cancer Research Program via the UCSF Helen Diller Family Comprehensive Cancer Center, George and Judy Marcus Award in Precision Medicine Innovation, UCSF Prostate Cancer Program, and Benioff Initiative for Prostate Cancer Research, UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation and UCSF Research Resource Fund.
Disclosure: Trey Ideker is a cofounder of Data4Cure, Inc., is on the scientific advisory board, and has an equity interest in the company; is on the scientific advisory board of Ideaya BioSciences, Inc., has an equity interest, and receives sponsored research funding from the company. J. Silvio Gutkind is a member of the board of scientific advisors for Vividion, Oncoceutics Pharmaceuticals and Domain Pharmaceuticals. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. Nevan Krogan’s laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. Krogan has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics, is a shareholder of Tenaya Therapeutics and has received stocks from Maze Therapeutics and Interline Therapeutics. Laura van’t Veer is a cofounder, stockholder and part-time employee of Agendia NV. Alan Ashworth is a cofounder of Tango Therapeutics, Azkarra Therapeutics, Ovibio Corporation, and Kytarro; is a consultant for SPARC, Bluestar, GenVivo, Earli, Cura, ProLynx, and GSK; is a member of the Scientific Advisory Board of Genentech and GLAdiator; receives grant and research support from SPARC and AstraZeneca; and holds patents on the use of PARP inhibitors held jointly with AstraZeneca, from which he has benefited financially and may do so in the future.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.