Computational Modeling Results in New Findings for Preeclampsia Patients
Study shows preeclampsia associated with low-oxygen microenvironment in uterus
Published Date
By:
- Kimberly Mann Bruch
Share This:
Article Content
According to the Centers for Disease Control and Prevention (CDC), preeclampsia, or pregnancy-related hypertension, occurs in roughly one in 25 pregnancies in the United States. The causes are unknown and childbirth is the only remedy, which can sometimes lead to adverse perinatal outcomes, such as preterm delivery. To better understand this serious pregnancy complication, which reduces blood supply to the fetus, researchers used Comet at the San Diego Supercomputer Center (SDSC) at UC San Diego to conduct cellular modeling to detail the differences between normal and preeclampsia placental tissue.
The researchers recently published findings from what they call their “disease-in-a dish” model in an article entitled Modeling Preeclampsia Using Human Induced Pluripotent Stem Cells in Scientific Reports.
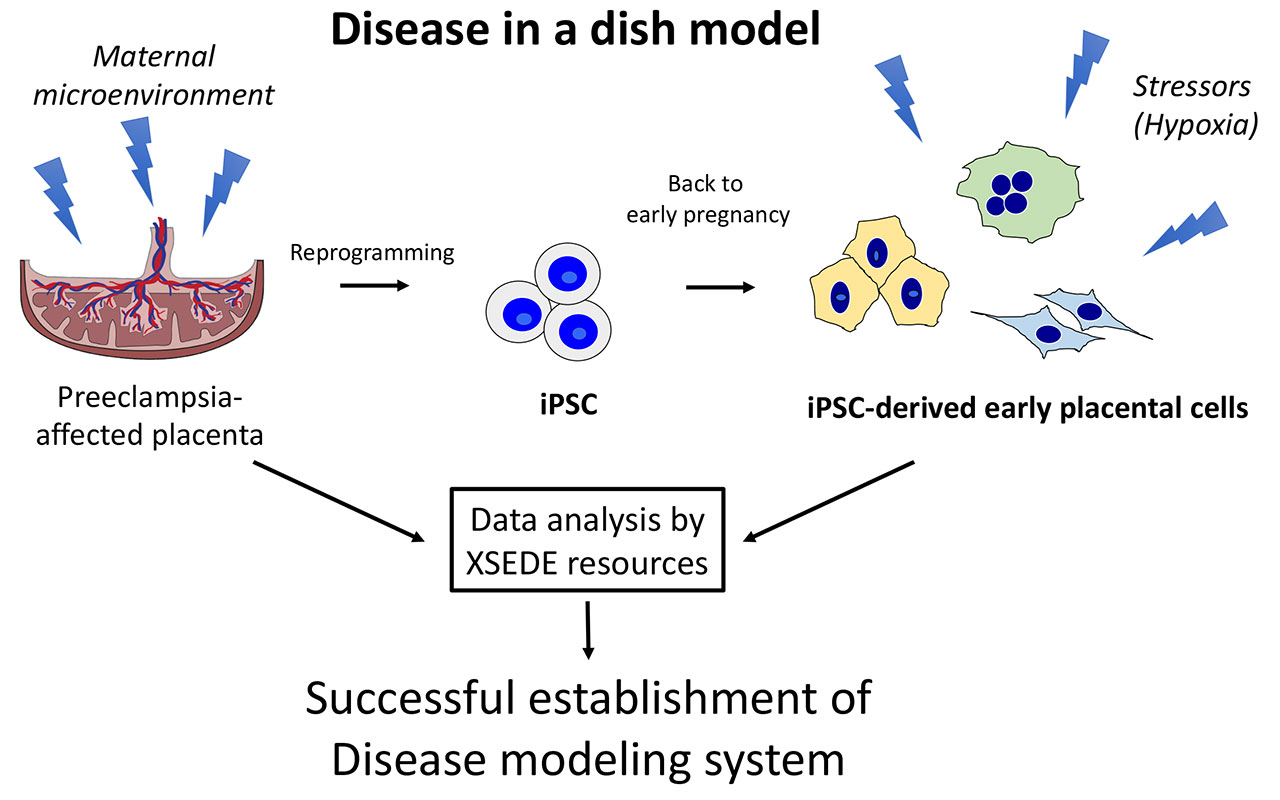
“We know this disease originates in early pregnancy, a period which is very difficult to study in an ongoing pregnancy, so we decided to take cells from the placenta at delivery and reprogram them into induced pluripotent stem cells, iPSC, which is like sending cells back in time,” said Mariko Horii, MD, principal investigator and assistant adjunct professor at UC San Diego School of Medicine. “The iPSC was then differentiated into cells resembling placental cells in early pregnancy and allowed our group to successfully recapitulate this disease in the dish—we were then able to examine the underlying pathophysiology of preeclampsia.”
The team’s analyses revealed that preeclampsia-associated cellular defects are partly secondary to an abnormal response to the maternal microenvironment. That is, preeclampsia is associated with a low-oxygen microenvironment in the uterus, and Comet-enabled calculations showed that the researchers’ model successfully re-created this scenario in vitro.
Why It’s Important
“Our group is inspired by all the suffering caused by pregnancy complications and are the result of a lack of understanding of the placenta, a vital organ for the baby during in-utero growth and development,” said Horii. “Because iPSC are patient-derived cells, they have been used for numerous cell-based modeling systems of multiple complex diseases such as Alzheimer’s, and our group applied this technology to study preeclampsia, with the hopes of advancing the science underlying pregnancy complications and thus promoting maternal and neonatal health.”
Credit: UC San Diego
This cell model system described by Horii not only allowed the researchers to better understand the fundamental concept of preeclampsia’s microenvironment, but also provided insight for future development of diagnostic tools and identification of potential medications. Further, Horii said that this iPSC-based model system can now be used to study other placenta-associated pregnancy disorders such as fetal growth restriction, miscarriage and preterm birth.
How SDSC Resources Helped
“Genomic data analysis is compute-intensive, and our access to Comet compute resources enabled us to create a cellular model for preeclampsia and discover differences between normal and preeclamptic placental tissue,” said Horii. “Using Comet allowed us to direct our resources toward modeling preeclampsia instead of diluting our efforts because we needed to determine details of compute resources.”
The research team first became acquainted with Comet and the National Science Foundation Extreme Science and Engineering Discovery Environment (XSEDE) in 2014, when colleagues received a start-up allocation at SDSC to help with their work that required complex genomic data to answer difficult biological questions. When they saw the success of their colleagues’ experience with XSEDE, they applied for and received their own start-up XSEDE allocation at SDSC in late 2014.
“Over the past seven years, SDSC’s Mahidhar Tatineni and Marty Kandes have helped us on countless occasions through the installation of necessary software and improving our data analysis scripts,” said Horii. “Access to XSEDE resources has enabled us to perform experiments at the genome level, whereas before we used to measure how pregnancy disorders affected a handful of genes. Now, with access to SDSC supercomputers, we are able to measure at the genome-wide level.”
What’s Next?
While the research team made great strides with this recent finding, they plan to study further the relationship between the maternal microenvironment and development of placental disease. Specifically, they’re developing a three-dimensional cellular model to study the maternal-fetal interface.
“Currently, model systems are in two-dimensional cultures with single-cell types, which are hard to study as the placenta consists of maternal and fetal cells with multiple cell types, such as placental cells (fetal origin), maternal immune cells and maternal endometrial cells,” explained Horii. “Combining these cell types together into a three-dimensional structure will lead to a better understanding of the more complex interactions and cell-to-cell signaling, which can then be applied to the disease setting to further understand pathophysiology. We look forward to our continued allocations from XSEDE to assist with this important future work.”
The research team was supported by California Institute for Regenerative Medicine (CIRM) Physician Scientist Award (RN3-06396), National Institutes of Health (NIH) (T32GM8806), NIH/National Institute of Child Health and Human Development (NICHD) (R01HD-089537 and K99 HD091452), CIRM Research and Training (TG2-01154), CIRM Bridges to Stem Cell Research Internship Program (EDUC2-08376), and CHA University School of Medicine. Computational data was generated at UC San Diego’s IGM Genomics Center utilizing an Illumina NovaSeq 6000 purchased with funding from NIH (t S10 OD026929) and used XSEDE allocation TG-MCB140074 on Comet for computational analysis, which is supported by NSF (ACI-1548562).
About SDSC
The San Diego Supercomputer Center (SDSC) is a leader and pioneer in high-performance and data-intensive computing, providing cyberinfrastructure resources, services, and expertise to the national research community, academia, and industry. Located on the UC San Diego campus, SDSC supports hundreds of multidisciplinary programs spanning a wide variety of domains, from astrophysics and earth sciences to disease research and drug discovery. SDSC’s newest NSF-funded supercomputer, Expanse, supports SDSC’s theme of “Computing without Boundaries” with a data-centric architecture, public cloud integration and state-of-the art GPUs for incorporating experimental facilities and edge computing.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



