By:
- Cynthia Dillon
Published Date
By:
- Cynthia Dillon
Share This:
Clearing the Course for Glycans in Development of Flu Drugs
Researchers demonstrate molecular binding mechanism that could change approach to designing influenza treatments
There is no hole-in-one drug treatment when it comes to the flu, but that doesn’t stop scientists from trying to sink one. Especially since as many as one in five Americans gets the flu. The reported estimated cost of this illness is $10 billion each year in medical expenses and another $16 billion in lost earnings in America alone, according to researchers at UC San Diego.

Rommie Amaro, Professor of Chemistry and Biochemistry, University of California, San Diego.
Teeing up on the science behind the flu virus is Rommie Amaro and J. Andrew McCammon, both professors of chemistry and biochemistry, and graduate student Christian Seitz. Together with co-workers Lorenzo Casalino, Robert Konecny and Gary Huber, they studied the effect of glycans—groups of sugar molecules—on the binding of antiviral drugs to viral neuraminidase. An enzyme found on the surface of flu viruses, neuraminidase enables the viruses to exit their diseased host cells and infect and replicate in new, previously healthy host cells. The glycans help to prevent large antibody molecules from binding to the enzyme.
The researchers’ findings, published in Biophysical Journal, likely apply more generally—including to the SARS-CoV-2 virus that causes COVID-19. Amaro will soon be releasing new findings about her latest research on the virus’ spike protein.
Zeroing in on glycans and binding sites
According to the scientists, influenza neuraminidase is the target for three FDA-approved influenza drugs in the U.S., but drug resistance and low drug effectiveness merit more drug development. Generally, however, drug developers do not include glycans in their development pipelines. They know glycans exist, but they have ignored glycans when designing new drugs without a basis for doing so and without evidence that glycans do not affect drug binding.
With their focus on glycans, the team thought it would be prudent to test the assumption about glycans in drug design relative to neuraminidase. Their results show that their proposed binding mechanism can help shed light on the complexity of the interplay between glycans and ligand binding.
“Traditionally, glycans have been difficult to study experimentally or theoretically due to a number of technological constraints, which are beginning to be lifted,” explained Seitz, first author of the study. “Because of this recent ‘emergence’ of glycans, we still have a lot to learn about them.”
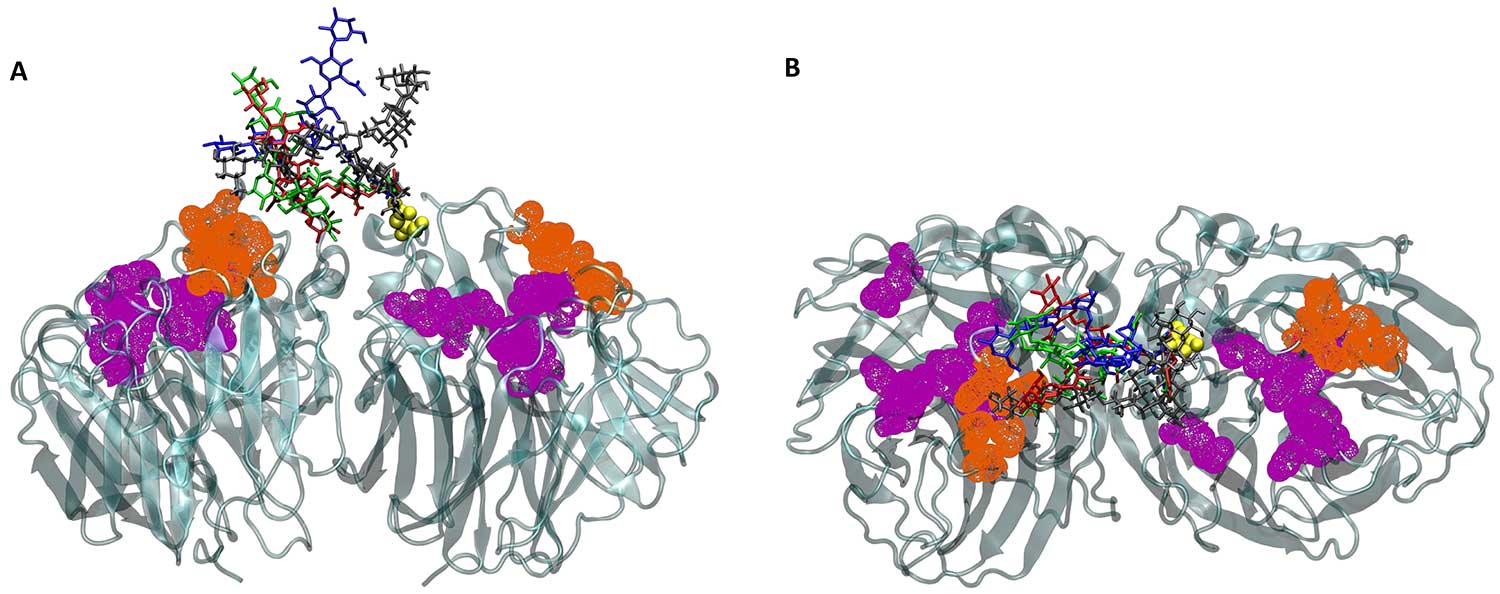
The superposition of four glycan conformations onto the static neuraminidase structure shows the conformational variability of the glycans, partially blocking binding site access. Metaphorically, the glycans are the rocks on the mini-golf course and the binding sites are the holes. The glycans are in blue, red, green and gray; the binding sites are in purple and orange, and neuraminidase is in teal. This representation is simplified to emphasize the relationship between the glycans and the binding sites, showing half of the neuraminidase structure and less than one-fifth of the glycans present in the full study. Figure by Christian Seitz, Amaro Lab and McCammon Lab, UC San Diego.
To better understand glycans in the context of this particular study, the team created all-atom in silico systems of influenza neuraminidase, consisting of four different glycan configurations and one glycan-free system. They observed a two- to eight-fold decrease in the rate of ligand binding to the primary binding site of neuraminidase After examining neuraminidase’s binding sites, the scientists noted that drugs prefer the primary binding site over the secondary binding site.
“Personally, I found two things to be quite surprising. Glycans are flexible and can reside very close to the drug binding sites, so I thought that the glycans would completely block drug binding. However, we found the glycans acted more like a screen or a curtain—things can get through, it will just take a bit longer,” said Seitz. “Secondly, there are two binding sites on influenza neuraminidase; one is the primary (catalytic) site needed for the viral replication cycle to continue, and the other, secondary site, is not well understood. Some previous studies had initially concluded that ligands would reach the secondary site significantly faster than the primary site.”
On par with progress
Seitz noted that McCammon was the first person to run a molecular dynamics study with a protein, and the Brownian dynamics software used in this study was developed in his lab. Additionally, Amaro is known as a world-leading expert in molecular dynamics virus simulations, and her virus work has recently been covered in The New York Times.
“Combining these rich basins of knowledge we are able to gain new insights into a global disease right here in San Diego,” said Seitz.
The graduate student likened the study to a mini-golf course. “We have the obvious goal of wanting to get the ball in the hole except, in our study, the golf ball is an influenza drug and the hole is the protein receptor the drug must find to kill the virus. One can often find large rocks on the greens acting as ‘gatekeepers’ to make it more difficult to get the ball in the hole. In our flu analogy, these rocks are the tiny sugars called glycans.”
Just as a groundskeeper can change the position of the rocks near the hole, glycans can change position on the protein. So, the researchers wanted to know if changing the position of the glycans would change how easy or difficult it is for the influenza drug to find its target.
“To start, we found common positions of these glycans. However, just as you would not change the position of the rocks on a mini-golf course and do one putt before declaring it easier or harder, we knew we would have to repeat this process many times (600 million, actually) to reach a statistically significant conclusion. Each Brownian dynamics trajectory in our study represented one putt on our mini-golf course, and we simply measured if the ball reached the hole,” Seitz explained.
The scientists found that changing the positions of the glycans did make it somewhat harder for the drug to reach the target, but not as much as expected. This means that drug developers do not need to account for glycans when designing new small-molecule drugs for influenza—something that was unclear before.
“For a long time, I thought this couldn't be true and ran numerous tests to disprove it, but all these tests consistently said the same thing, that most small ligands are able to evade the glycans and bind to the enzyme,” Seitz said. “Thus, this work is one small step in helping to ameliorate the yearly human and economic cost in our nation and our world. This is paid for by our own taxes so each of us has made a tiny contribution to this progress.”
This research was supported in part by grants from the National Institutes of Health, USA (NIH grant nos. T32EB009380 and GM031749 and the National Science Foundation Graduate Research Fellowship Program (grant no. DGE-1650112). The researchers used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (grant no. ACI-1548562) and the XSEDE Comet at the San Diego Supercomputer Center (allocation csd373).
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



