Brain Organoids Mimic Head Size Changes Associated with Type of Autism
Stem cell models derived from people with specific genomic variation recapitulate aspects of their autism spectrum disorder, providing a valuable model to study the condition and look for therapeutic interventions
By:
- Heather Buschman, PhD
Published Date
By:
- Heather Buschman, PhD
Share This:
Article Content
Variations in the 16p11.2 region of the genome are associated with autism spectrum disorder. While people with genetic deletions in this region have larger heads (macrocephaly) and people with genetic duplications have smaller heads (microcephaly), both variation types affect brain development and function.
To study the effects of these variations and search for ways to minimize their impact, University of California San Diego School of Medicine researchers are using brain organoids — tiny, 3D cellular models generated in the lab from people with 16p11.2 variations.
The organoids, described in a paper publishing August 25, 2021 in Molecular Psychiatry, mimicked the differences in brain size seen in people. They also revealed new information about the molecular mechanisms that malfunction when the 16p11.2 region of the genome is disrupted, providing new opportunities for potential therapeutic intervention.
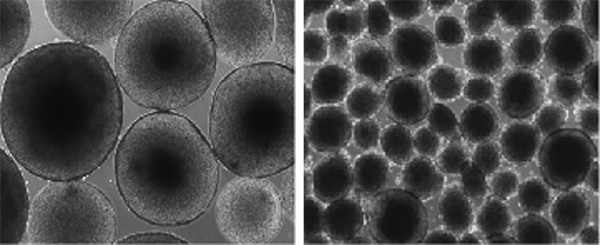
Variations in the 16p11.2 region of the genome are associated with autism spectrum disorder and changes in head size. Brain organoids grown in the lab with a 16p11.2 deletion demonstrate macrocephaly (larger size, left), while 16p11.2 duplication demonstrates microcephaly (smaller size, right).
“Because our organoids recapitulate the head size of the patients, that tells us this can be a useful model,” said senior author Lilia Iakoucheva, PhD, associate professor of psychiatry at UC San Diego School of Medicine. “And we need better models to study autism spectrum disorder, especially during fetal development.”
Iakoucheva led the study with Alysson Muotri, PhD, professor of pediatrics and cellular and molecular medicine at UC San Diego School of Medicine.
The brain organoids were created using induced pluripotent stem cells derived from people who have 16p11.2 genomic variations — three people with deletions, three with duplications and three non-variant controls. Researchers obtained a skin sample from each person, gave the skin cells a molecular cocktail that converted them to stem cells, then treated the stem cells in a way that coaxed them into becoming brain cells, preserving each patient’s unique genetic background.
The organoids revealed that RhoA — a protein that plays a big part in many basic cellular functions, such as development and movement — is more active in both 16p11.2-deleted and 16p11.2-duplicated organoids than it is in organoids without these variations. Over-active RhoA led to a slowdown in neuronal migration, the process by which brain cells get to where they need to be for normal fetal development and function in adulthood.
When the team inhibited RhoA in the autism-like organoids, neuronal migration was restored to levels seen in the control organoids.
“Our work opens the possibility to therapeutically manipulate the RhoA pathway,” said Muotri, who is also director of the UC San Diego Stem Cell Program and a member of the Sanford Consortium for Regenerative Medicine. “The same pathway may be also damaged in other individuals with autism spectrum disorder who have macrocephaly or microcephaly. Considering this, we can potentially help millions of patients.”
Organoids aren’t perfect reproductions of the brain. They lack connections to other organ systems, such as blood vessels, and so don’t encapsulate full human biology. In addition, therapeutics tested on brain organoids are added directly. They don’t need to get across the blood-brain barrier, specialized blood vessels that keep the brain largely free of microbes and toxins.
The team plans to further test RhoA inhibitors in a mouse model with 16p11.2 variations or over-active RhoA for their ability to reverse defects associated with autism spectrum disorder.
Co-authors include: Jorge Urresti, Pan Zhang, Patricia Moran-Losada, Priscilla D. Negraes, Cleber A. Trujillo, Danny Antaki, Megha Amar, Kevin Chau, Akula Bala Pramod, Leon Tejwani, Sarah Romero, and Jonathan Sebat, all at UC San Diego; Nam-Kyung Yu, Jolene Diedrich, and John R. Yates III, Scripps Research.
Funding for this research came, in part, from the Simons Foundation for Autism Research (grant 345469) and National Institutes of Health (grants MH109885, MH108528, MH105524, MH104766, MH100175, MH119746, P30CA023100, NS047101).
Disclosure: Alysson R. Muotri is a co-founder and has equity interest in TISMOO, a company dedicated to genetic analysis and brain organoid modeling focusing on therapeutic applications customized for autism spectrum disorder and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



