Ambitious Project to Understand Cellular Evolution Underway
Research seeks to understand the “rules” of cellular architecture for eukaryotic cells
Published Date
By:
- Cynthia Dillon
Share This:
Article Content
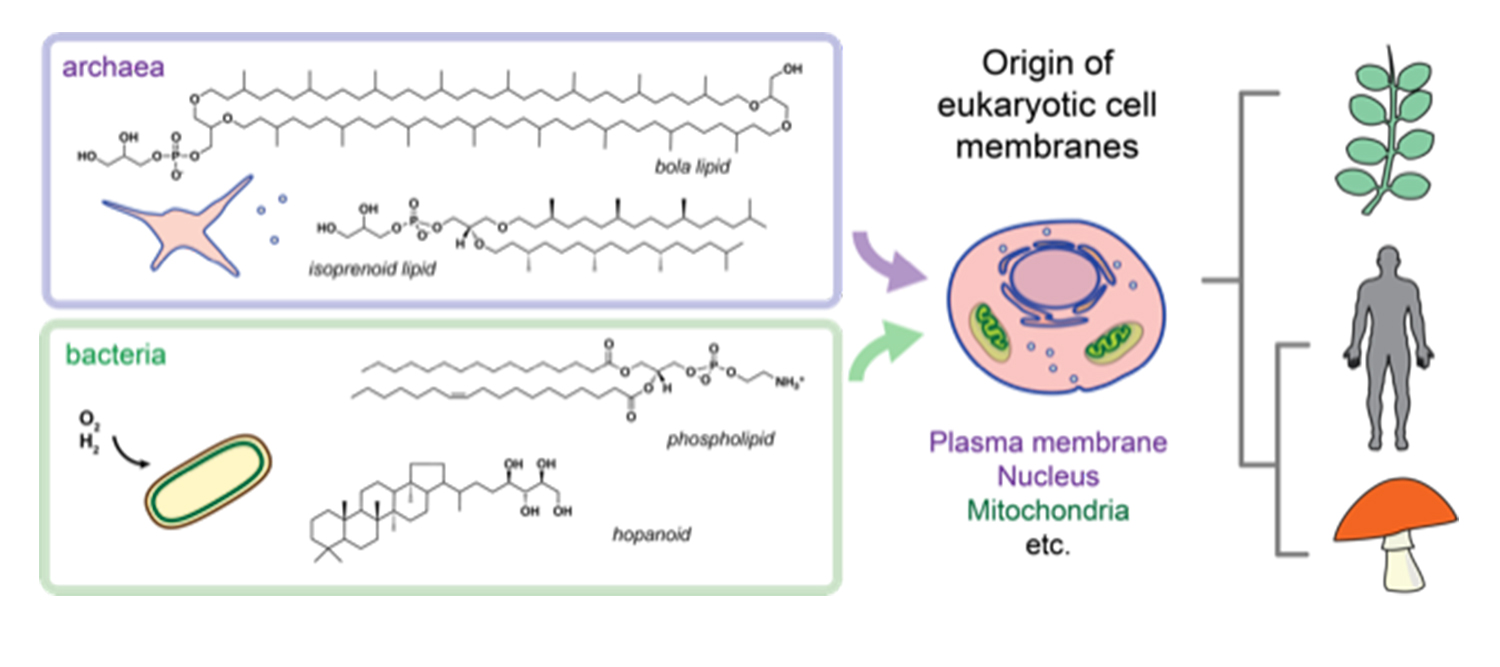
The two main lineages of prokaryotic cells--archaea and bacteria—together form the basis of all eukaryotic cells that compose animals, plants and fungi, among others. UC San Diego researchers are trying to uncover how the different lipid chemistries and membrane structures in these lineages of life first evolved. Credit: Itay Budin
The study of the early evolution of life is changing. With nearly $900,000 in new funding from the Gordon and Betty Moore Foundation (through the Moore-Simons Project on the Origin of the Eukaryotic Cell supported by the Moore Foundation and the Simons Foundation), researchers at UC San Diego are working to understand how the cellular precursors to plants and animals evolved 2 billion years ago—a fundamental evolutionary event that completely transformed life on Earth. The research funding contributes to the Campaign for UC San Diego.
Cells are the fundamental unit of life, and before more complex plant, animal and fungi cells emerged, there were single-celled organisms called prokaryotes. These microbes had no nucleus or organelles (separate membrane-bound compartments with specific functions in complex cells). When prokaryote cells evolved into eukaryotic cells with multiple internal organelles and the associated machinery for transport between them, it led to a tremendous increase in the complexity of cellular life.
According to Itay Budin, the principal investigator on the project, it is generally accepted that eukaryotic cells resulted from the merging of archaea and bacteria, the two main lineages of prokaryotic life, a concept promoted in 1967 by Lynn Margulis, an American biologist who modernized the concept of how life developed on Earth. But scientists still do not understand the exact biochemical and environmental processes that allowed for this transition.
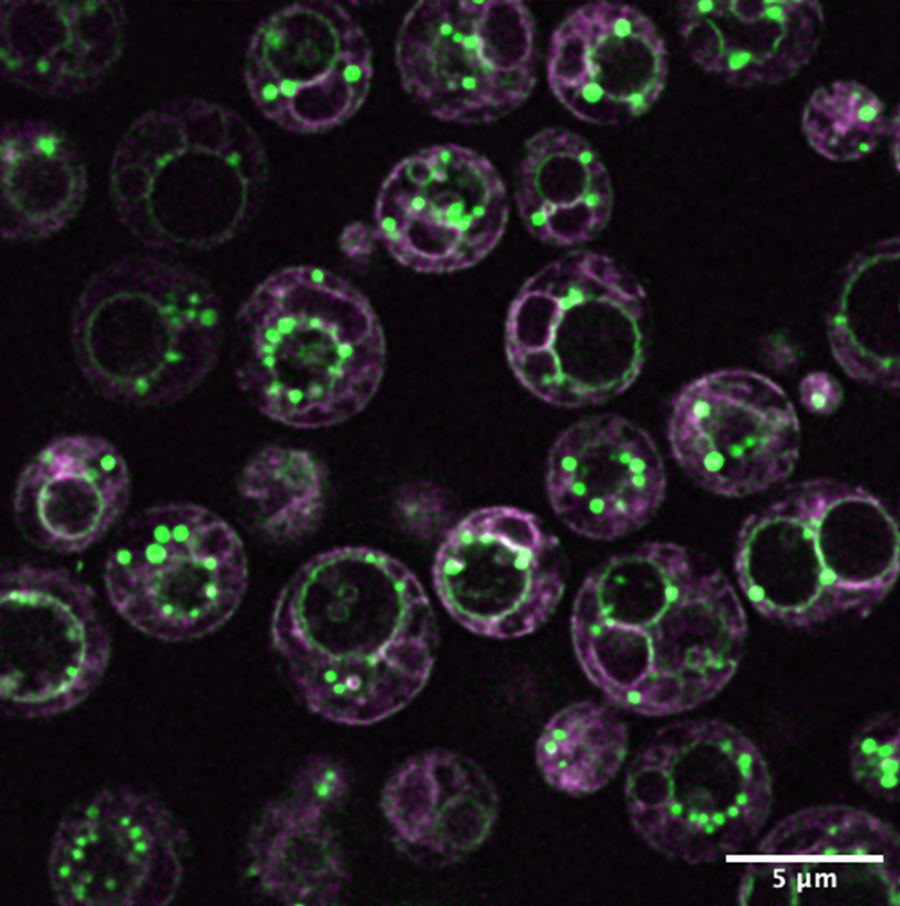
The multiple membranes of yeast cells, imaged using lipid-associated fluorophores. The project uses yeast as a model system to study the distinct membrane compartments found in all eukaryotic cells. Credit: Itay Budin
As the research seeks to understand the “rules” of cellular architecture for eukaryotic cells—carrying implications for how human cells function in health and disease, as well as lead to the development of imaging technologies that can be broadly applied in the biomedical sciences—it will focus on lipids (fat molecules) as the starting point to understand the evolution of eukaryotic cells, since their organelles are defined by membranes made from lipids.
“Interestingly, archaea and bacteria, which joined to form the first eukaryotes, have completely distinct types of lipid metabolism—they make fundamentally different membrane-building blocks,” explained Budin, an assistant professor in the Department of Chemistry and Biochemistry. “We want to understand how these molecular pathways mixed, generated new lipid species that are unique to eukaryotes (e.g., cholesterol) and formed the key membranes that make up all eukaryotic cells, including our own.”
Budin said that currently scientists can’t answer questions like, “What conditions are needed for complex cells to arise?” or “How likely are these to occur on other planets?” He said that studying evolution is broadly important because it helps us understand how organisms can adapt to changing environments on our planet. For example, he noted efforts to understand the interplay between oxygen and lipid chemistry, which is important for understanding how marine organisms might respond to deoxygenation in the oceans due to climate change.
“Key steps in this pathway require oxygen, so reconstructing their ancestral enzymes is a way to gain insight on how biochemistry evolved in that environment,” he said.
The research will combine wet lab work with modeling and computational approaches, including the mining of genome sequences across life to generate “primordial” enzymes involved in lipid biosynthesis that can be purified or introduced into yeast.
“Because finding fossil records of single-cell life is a challenge, we need to turn to laboratory or synthetic methods to understand this evolutionary process,” said Budin. “This involves devising experiments and simulations in the lab that we can use to reconstruct potential cellular transition states to understand why and how natural selection occurred in this way, and also shed light on the potential constraints that eukaryogenesis (the emergence of eukaryotic cells) placed on the environment in which it occurred.”
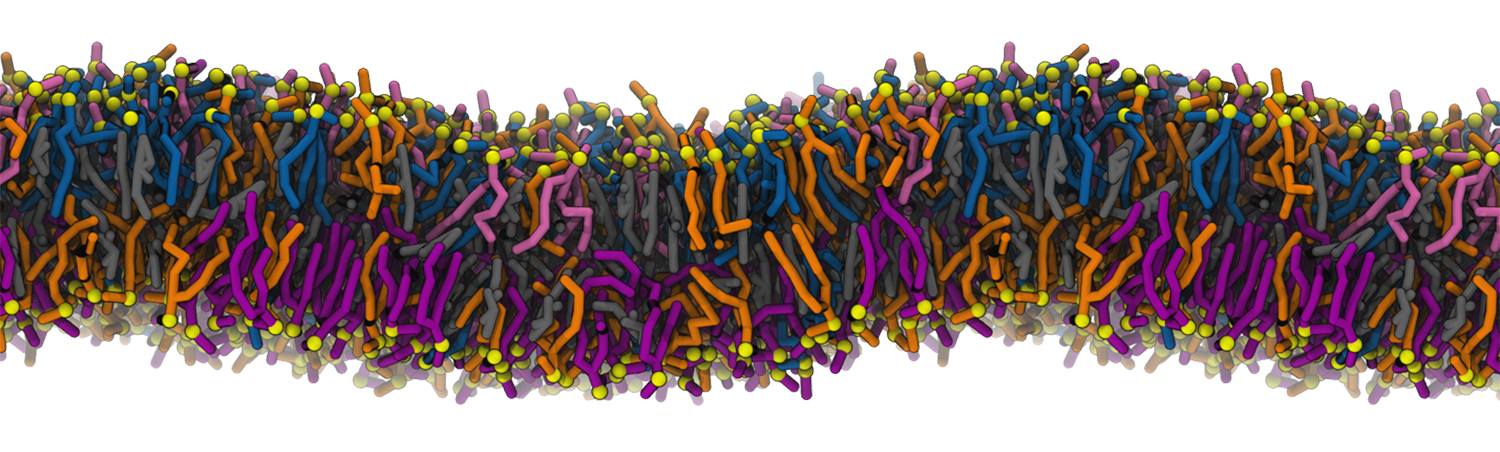
A molecular dynamics simulation of a cell membrane composed of individual lipid molecules. Simulations by the Rangamani Group will be used to model how changes in lipid chemistry affected key membrane properties during cellular evolution. Credit: Padmini Rangamani
The evolving public health situation with COVID-19 will also necessitate the use of digital forms of communications to aid in the research collaboration, which includes Mark Ellisman of the UC San Diego School of Medicine and Padmini Rangamani of the Jacobs School of Engineering. Patrick Shih from UC Davis’ Department of Plant Biology, College of Biological Sciences, is also part of the team.
Assisting with the study will be UC San Diego postdoctoral scholars Yi-Ting Tsai and Will Moore (no relation to the foundation). Tsai will be altering the lipids of mitochondrial membranes—the powerhouse of the cell—to determine how they influence the structure of this key eukaryotic membrane. Moore will be collaborating with Ellisman’s lab on developing new techniques to image membranes with electron microscopy, which is the same technology being applied to understanding how cells function in our brain and other tissues, and mechanical modeling to understand membrane processes central to eukaryotic cells.
“The team-building aspect of the project will certainly be different from what we first envisioned—way more Zoom meetings, way less traveling for the moment,” said Budin. “On the positive side, I think it will make it easier for the trainees involved in the project in California to stay connected and share data. UC San Diego is such a fantastic place for collaboration, which is key for taking on this highly ambitious study.”
Private support, like this funding from the Gordon and Betty Moore Foundation and the Simons Foundation, contribute to the Campaign for UC San Diego—a university-wide comprehensive fundraising effort concluding in 2022. Alongside UC San Diego’s philanthropic partners, the university is continuing its nontraditional path toward revolutionary ideas, unexpected answers, lifesaving discoveries and planet-changing impact. Visit the website to learn more about supporting the UC San Diego Division of Physical Sciences.
Share This:
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



