We Might Not Know Half of What’s in Our Cells, New AI Technique Reveals
Artificial intelligence-based technique reveals previously unknown cell components that may provide new clues to human development and disease
Published Date
By:
- Heather Buschman
Share This:
Article Content
Most human diseases can be traced to malfunctioning parts of a cell — a tumor is able to grow because a gene wasn’t accurately translated into a particular protein or a metabolic disease arises because mitochondria aren’t firing properly, for example. But to understand what parts of a cell can go wrong in a disease, scientists first need to have a complete list of parts.
By combining microscopy, biochemistry techniques and artificial intelligence, researchers at University of California San Diego School of Medicine and collaborators have taken what they think may turn out to be a significant leap forward in the understanding of human cells.
The technique, known as Multi-Scale Integrated Cell (MuSIC), is described November 24, 2021, in Nature.
“If you imagine a cell, you probably picture the colorful diagram in your cell biology textbook, with mitochondria, endoplasmic reticulum and nucleus. But is that the whole story? Definitely not,” said Trey Ideker, PhD, professor at UC San Diego School of Medicine and Moores Cancer Center. “Scientists have long realized there’s more that we don’t know than we know, but now we finally have a way to look deeper.”
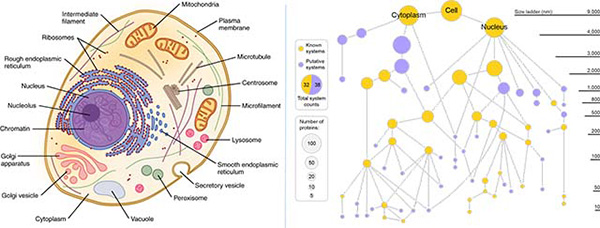
Left: Classic textbook cell diagrams imply all parts are clearly visible and defined. (Credit: OpenStax/Wikimedia). Right: A new cell map generated by MuSIC technic reveals many novel components. Gold nodes represent known cell components, purple nodes represent new components. The size of node reflects number of distinct proteins in that component.
Ideker led the study with Emma Lundberg, PhD, of KTH Royal Institute of Technology in Stockholm, Sweden and Stanford University.
In the pilot study, MuSIC revealed approximately 70 components contained within a human kidney cell line, half of which had never been seen before. In one example, the researchers spotted a group of proteins forming an unfamiliar structure. Working with UC San Diego colleague Gene Yeo, PhD, they eventually determined the structure to be a new complex of proteins that binds RNA. The complex is likely involved in splicing, an important cellular event that enables the translation of genes to proteins, and helps determine which genes are activated at which times.
The insides of cells — and the many proteins found there — are typically studied using one of two techniques: microscope imaging or biophysical association. With imaging, researchers add florescent tags of various colors to proteins of interest and track their movements and associations across the microscope’s field of view. To look at biophysical associations, researchers might use an antibody specific to a protein to pull it out of the cell and see what else is attached to it.
The team has been interested in mapping the inner workings of cells for many years. What’s different about MuSIC is the use of deep learning to map the cell directly from cellular microscopy images.
“The combination of these technologies is unique and powerful because it’s the first time measurements at vastly different scales have been brought together,” said study first author Yue Qin, a Bioinformatics and Systems Biology graduate student in Ideker’s lab.
Microscopes allow scientists to see down to the level of a single micron, about the size of some organelles, such as mitochondria. Smaller elements, such as individual proteins and protein complexes, can’t be seen through a microscope. Biochemistry techniques, which start with a single protein, allow scientists to get down to the nanometer scale. (A nanometer is one-billionth of a meter, or 1/1,000th of a micron.)
“But how do you bridge that gap from nanometer to micron scale? That has long been a big hurdle in the biological sciences,” said Ideker, who is also founder of the UC Cancer Cell Map Initiative and the UC San Diego Center for Computational Biology and Bioinformatics. “Turns out you can do it with artificial intelligence — looking at data from multiple sources and asking the system to assemble it into a model of a cell.”
The team trained the MuSIC artificial intelligence platform to look at all the data and construct a model of the cell. The system doesn’t yet map the cell contents to specific locations, like a textbook diagram, in part because their locations aren’t necessarily fixed. Instead, component locations are fluid and change depending on cell type and situation.
Ideker noted this was a pilot study to test MuSIC. They’ve only looked at 661 proteins and one cell type.
“The clear next step is to blow through the entire human cell,” Ideker said, “and then move to different cell types, people and species. Eventually we might be able to better understand the molecular basis of many diseases by comparing what’s different between healthy and diseased cells.”
Co-authors include: Maya L. Gosztyla, Marcus R. Kelly, Steven M. Blue, Fan Zheng, Michael Chen, Leah V. Schaffer, Katherine Licon, John J. Lee, Sophie N. Liu, Erica Silva, Jisoo Park, Adriana Pitea, Jason F. Kreisberg, UC San Diego; Edward L. Huttlin, Laura Pontano Vaites, Tian Zhang, Steven P. Gygi, J. Wade Harper, Harvard Medical School; Casper F. Winsnes, Anna Bäckström, Wei Ouyang, KTH Royal Institute of Technology; Ludivine Wacheul, Denis L. J. Lafontaine, Université Libre de Bruxelles; and Jianzhu Ma, Peking University.
Funding for this research came, in part, from the National Institutes of Health (grants U54CA209891, U01MH115747, F99CA264422, P41GM103504, R01HG009979, U24HG006673, U41HG009889, R01HL137223, R01HG004659, R50CA243885), Google Ventures, Erling-Persson Family Foundation, Knut and Alice Wallenberg Foundation (grant 2016.0204), Swedish Research Council (grant 2017-05327), Belgian Fonds de la Recherche Scientifique, Université Libre de Bruxelles, European Joint Programme on Rare Diseases, Région Wallonne, Internationale Brachet Stiftung, and Epitran COST action (grant CA16120).
Disclosures: Trey Ideker is co-founder of, on the Scientific Advisory Board and has an equity interest in Data4Cure, Inc. Ideker is also on the Scientific Advisory Board, has an equity interest in and receives sponsored research funding from Ideaya BioSciences, Inc. Gene Yeo is a co-founder, member of the Board of Directors, on the Scientific Advisory Board, an equity holder and a paid consultant for Locanabio and Eclipse BioInnovations. Yeo is also a visiting professor at the National University of Singapore. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies. Emma Lundberg is on the Scientific Advisory Boards of and has equity interests in Cartography Biosciences, Nautilus Biotechnology and Interline Therapeutics. J. Wade Harper is a co-founder of, on the Scientific Advisory Board and has an equity interest in Caraway Therapeutics. Harper is also Founding Scientific Advisor for Interline Therapeutics.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.



